













| General Information: |
|
| Name(s) found: |
HOM6 /
YJR139C
[SGD]
|
| Description(s) found:
Found 39 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 359 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
response to drug
[IMP methionine metabolic process [TAS threonine metabolic process [TAS homoserine biosynthetic process [TAS] |
| Molecular Function: |
homoserine dehydrogenase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTKVVNVAV IGAGVVGSAF LDQLLAMKST ITYNLVLLAE AERSLISKDF SPLNVGSDWK 60
61 AALAASTTKT LPLDDLIAHL KTSPKPVILV DNTSSAYIAG FYTKFVENGI SIATPNKKAF 120
121 SSDLATWKAL FSNKPTNGFV YHEATVGAGL PIISFLREII QTGDEVEKIE GIFSGTLSYI 180
181 FNEFSTSQAN DVKFSDVVKV AKKLGYTEPD PRDDLNGLDV ARKVTIVGRI SGVEVESPTS 240
241 FPVQSLIPKP LESVKSADEF LEKLSDYDKD LTQLKKEAAT ENKVLRFIGK VDVATKSVSV 300
301 GIEKYDYSHP FASLKGSDNV ISIKTKRYTN PVVIQGAGAG AAVTAAGVLG DVIKIAQRL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
AHA1 BUD3 CNS1 CPR6 DIG2 DNM1 DSF1, YNR073C FRT2 GPA2 HOM6 HSC82 HSP104 HSP82 MBB1 MRPL50 PAM1 PPT1 QNS1 RMD8 RTS1 SMT3 STI1 STP4 TDH1 UTP20 YBL104C |
| View Details | Ho Y, et al. (2002) |
|
ECM29 GYP5 HOM6 PHO84 RVS167 UBI4 UBP7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
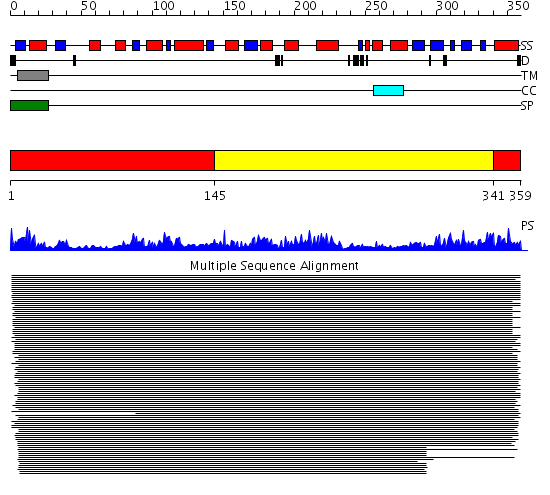
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..144] [341..359] |
10080.69897 | Homoserine dehydrogenase | |
| 2 | View Details | [145..340] | 10080.69897 | Homoserine dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|