













| General Information: |
|
| Name(s) found: |
SPA2 /
YLL021W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1466 amino acids |
Gene Ontology: |
|
| Cellular Component: |
polarisome
[TAS incipient cellular bud site [IDA cellular bud tip [IDA cellular bud neck [IDA mating projection tip [IDA |
| Biological Process: |
bipolar cellular bud site selection
[TAS regulation of initiation of mating projection growth [IMP pseudohyphal growth [IMP actin filament organization [IPI regulation of termination of mating projection growth [IMP Rho protein signal transduction [IPI |
| Molecular Function: |
cytoskeletal regulatory protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTSSEVSLA HHRDIFHYYV SLKTFFEVTG ENRDRSNSTR AQKARAKLLK LSSSQFYELS 60
61 TDVSDELQRR IGEDANQPDY LLPKANFHMK RNQARQKLAN LSQTRFNDLL DDILFEIKRR 120
121 GFDKDLDAPR PPLPQPMKQE VSKDSDDTAR TSTNSSSVTQ VAPNVSVQPS LVIPKMASID 180
181 WSSEEEEEEQ VKEKPNEPEG KQTSMDEKKE AKPALNPIVT DSDLPDSQVL ARDITSMART 240
241 PTTTHKNYWD VNDSPIIKVD KDIDNEKGPE QLKSPEVQRA ENNNPNSEME DKVKELTDLN 300
301 SDLHLQIEDL NAKLASLTSE KEKEKKEEKE EKEKEKNLKI NYTIDESFQK ELLSLNSQIG 360
361 ELSIENENLK QKISEFELHQ KKNDNHNDLK ITDGFISKYS SADGLIPAQY ILNANNLIIQ 420
421 FTTRLSAVPI GDSTAISHQI GEELFQILSQ LSNLISQLLL SADLLQYKDQ VILLKASLSH 480
481 AITSIRYFSV YGPVLIPKIT VQAAVSEVCF AMCNLIDSAK IKSDSNGEST TSNEGNRQVL 540
541 EYSSPTATTP MTPTFPSTSG INMKKGFINP RKPASFLNDV EEEESPVKPL KITQKAINSP 600
601 IIRPSSSNGV PTTSRKPSGT GLFSLMIDSS IAKNSSHKED NDKYVSPIKA VTSASNSASS 660
661 NISEIPKLTL PPQAKIGTVI PPSENQVPNI KIENTEEDNK RSDITNEISV KPTSSIADKL 720
721 KQFEQSSEKK SSPKENPIAK EEMDSKPKLS NKFITSMNDV STDDSSSDGN ENDDADDDDD 780
781 FTYMALKQTM KREGSKIEKN NDSKLPANIV ELDLHESPES VKIESPESIK EITSSEMSSE 840
841 MPSSSLPKRL VEDVEPSEMP EKGASVESVR KKNFQEPLGN VESPDMTQKV KSLGMTGKAV 900
901 GPESDSRVES PGMTGQIKSL NMAGKVVGPE ADSRVESPGM KEQIKSLGMT GKITAQESIK 960
961 SPEAARKLAS SGEVDKIESP RMVRESESLE AVGNTIPSNM TVKMESPNLK GNTVSEPQEI1020
1021 RRDIASSEPI ENVDPPKVLK KIVFPKAVNR TGSPKSVEKT PSSATLKKSG LPEPNSQIVS1080
1081 PELAKNSPLA PIKKNVELRE TNKPHTETIT SVEPTNKDAN TSWRDADLNR TIKREEEDED1140
1141 FDRVNHNIQI TGAYTKTGKI DYHKIPVDRK AKSEAEVHTS EEDIDESNNV NGKRADAQIH1200
1201 ITERKHAFVN PTENSQVKKT SHSPFLNSKP VQYENSESNG GINNHIKIKN TGETTAHDEK1260
1261 HYSDDDDSSY QFVPMKHEEQ EQEQNRSEEE ESEDDDEEEE DSDFDVDTFD IENPDNTLSE1320
1321 LLLYLEHQTM DVISTIQSLL TSIKKPQVTK GNLRGESNAI NQVIGQMVDA TSISMEQSRN1380
1381 ANLKKHGDWV VQSLRDCSRR MTILCQLTGD GILAKEKSDQ DYADKNFKQR LAGIAFDVAK1440
1441 CTKELVKTVE EASLKDEINY LNSKLK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
PEA2 SPA2 |
| View Details | Krogan NJ, et al. (2006) |
|
BEM2 PEA2 SPA2 |
| View Details | Gavin AC, et al. (2006) |
|
DBP9 PSE1 SLX9 SPA2 ULP1 YMR310C |
| View Details | Gavin AC, et al. (2002) |

|
BLM10 DAM1 DBP9 ECM29 EST3 GFA1 INO4 KAP95 LYS12 MDS3 NUD1 PDA1 PDB1 PRE10 PRE2 PRE3 PRE4 PRE5 PRE6 PRE8 PRE9 PSE1 PUP3 RGR1 RPT3 RPT5 RRP12 SCL1 SLX9 SPA2 SRP1 ULP1 YFL006W YMR310C YRA1 |
| View Details | Qiu et al. (2008) |
|
BEM1 BNI1 BNR1 BOI1 BOI2 BUD6 CRN1 HOF1 IQG1 LAS17 MLC1 MYO2 PAN1 PEA2 PXL1 RVS161 SLA1 SLA2 SPA2 SPH1 SRV2 STU2 VRP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Drees BL, et al. (2001) | ||
| View Screen | 2 | Drees BL, et al. (2001) |
New Feature: Upload Your Own Microscopy Data
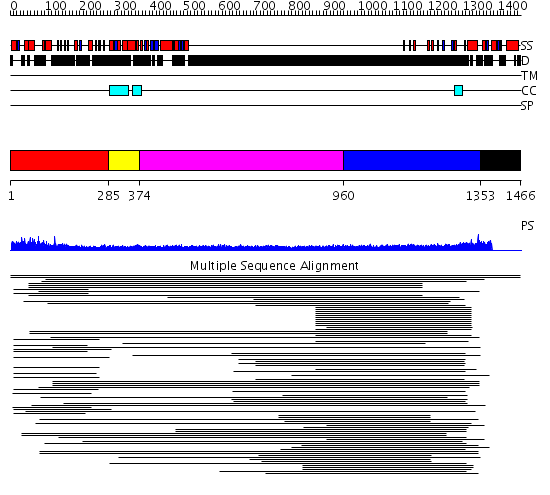
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..284] | 7.060967 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [285..373] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [374..959] | 1.163997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [960..1352] | 24.221997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1353..1466] | N/A | Confident ab initio structure predictions are available. | |
| 6 | View Details | [825..947] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [948..1100] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1101..1353] | 2.403996 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1354..1466] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)