













| General Information: |
|
| Name(s) found: |
SSA3 /
YBL075C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 649 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
SRP-dependent cotranslational protein targeting to membrane, translocation
[IMP protein folding [IGI response to stress [IMP |
| Molecular Function: |
ATPase activity
[IGI unfolded protein binding [IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRAVGIDLG TTYSCVAHFS NDRVEIIAND QGNRTTPSYV AFTDTERLIG DAAKNQAAIN 60
61 PHNTVFDAKR LIGRKFDDPE VTTDAKHFPF KVISRDGKPV VQVEYKGETK TFTPEEISSM 120
121 VLSKMKETAE NYLGTTVNDA VVTVPAYFND SQRQATKDAG TIAGMNVLRI INEPTAAAIA 180
181 YGLDKKGRAE HNVLIFDLGG GTFDVSLLSI DEGVFEVKAT AGDTHLGGED FDNRLVNHLA 240
241 TEFKRKTKKD ISNNQRSLRR LRTAAERAKR ALSSSSQTSI EIDSLFEGMD FYTSLTRARF 300
301 EELCADLFRS TLEPVEKVLK DSKLDKSQID EIVLVGGSTR IPKIQKLVSD FFNGKEPNRS 360
361 INPDEAVAYG AAVQAAILTG DQSTKTQDLL LLDVAPLSLG IETAGGIMTK LIPRNSTIPT 420
421 KKSETFSTYA DNQPGVLIQV FEGERTRTKD NNLLGKFELS GIPPAPRGVP QIDVTFDIDA 480
481 NGILNVSALE KGTGKSNKIT ITNDKGRLSK DDIDRMVSEA EKYRADDERE AERVQAKNQL 540
541 ESYAFTLKNT INEASFKEKV GEDDAKRLET ASQETIDWLD ASQAASTDEY KDRQKELEGI 600
601 ANPIMTKFYG AGAGAGPGAG ESGGFPGSMP NSGATGGGED TGPTVEEVD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CMR2 POR1 SSA3 SSB2 |
| View Details | Krogan NJ, et al. (2006) |
|
APP1 CST6 HMO1 MCM10 MIG1 SSA3 YRM1 |
| View Details | Gavin AC, et al. (2002) |
|
ACT1 AIM31 AVO1 BEM2 CEG1 CET1 CFT1 CFT2 CKA1 CLP1 CLU1 CTR9 ENO2 FIP1 GLC7 HHF1, HHF2 HTA1 LSM12 MPE1 NIP1 NOP1 PAP1 PCF11 PDB1 PFK1 PFS2 PRP43 PTA1 PTI1 REF2 RNA14 RNA15 RPG1 RSA3 SEC13 SEC31 SPT16 SSA3 SSA4 SSU72 SYC1 TAF6 TIF4632 TOP2 TYE7 VID24 VMR1 VPS1 VPS53 YCL046W YSH1 YTH1 |
The following runs contain data for this protein:
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
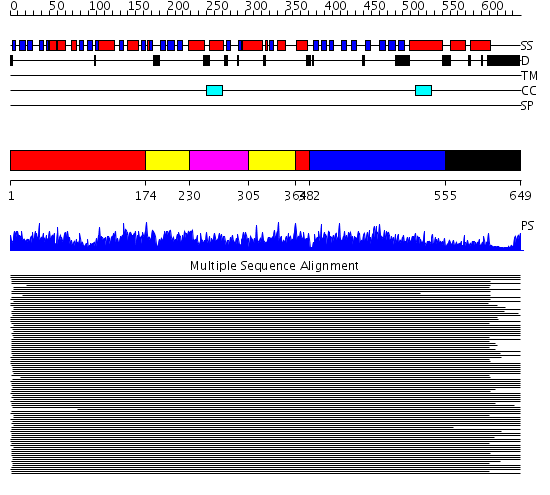
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..173] [364..381] |
2750.0 | Heat shock protein 70kDa, ATPase fragment | |
| 2 | View Details | [174..229] [305..363] |
2750.0 | Heat shock protein 70kDa, ATPase fragment | |
| 3 | View Details | [230..304] | 2750.0 | Heat shock protein 70kDa, ATPase fragment | |
| 4 | View Details | [382..554] | 739.37734 | DnaK | |
| 5 | View Details | [555..649] | 7.52 | DnaK |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)