













| General Information: |
|
| Name(s) found: |
CMR2 /
YOR093C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1648 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDFSIPPTLP LDLQSRLNEL IQDYKDENLT RKGYETKRKQ LLDKFEISQM RPYTPLRSPN 60
61 SRKSKHLHRR NTSLASSITS LPNSIDRRHS IYRVTTINST SANNTPRRRS KRYTASLQSS 120
121 LPGSSDENGS VKDAVYNPMI PLLPRHTGAE NTSSGDSAMT DSLPLILRGR FEHYDGQTAM 180
181 ISINSKGKET FITWDKLYLK AERVAHELNK SHLYKMDKIL LWYNKNDVIE FTIALLGCFI 240
241 SGMAAVPVSF ETYSLREILE IIKVTNSKFV LISNACHRQL DNLYSSSNHS KVKLVKNDVF 300
301 QQIKFVKTDD LGTYTKAKKT SPTFDIPNIS YIEFTRTPLG RLSGVVMKHN ILINQFETMT 360
361 KILNSRSMPH WKQKSQSIRK PFHKKIMATN SRFVILNSLD PTRSTGLIMG VLFNLFTGNL 420
421 MISIDSSILQ RPGGYENIID KFRADILLND QLQLKQVVIN YLENPESAFS KKHKIDFSCI 480
481 KSCLTSCTTI DTDVSEMVVH KWLKNLGCID APFCYSPMLT LLDFGGIFIS IRDQLGNLEN 540
541 FPIHNSKLRL QNELFINREK LKLNEVECSI TAMINSSSSF KDYLKLETFG FPIPDITLCV 600
601 VNPDTNTLVQ DLTVGEIWIS SNHITDEFYQ MDKVNEFVFK AKLNYSEMFS WAKYEMPTNE 660
661 KSQAVTEQLD TILNICPANT YFMRTKLMGF VHNGKIYVLS LIEDMFLQNR LIRLPNWAHT 720
721 SNLLYAKKGN QSAQPKGNTG AESTKAIDIS SLSGETSSGY KRVVESHYLQ QITETVVRTV 780
781 NTVFEVAAFE LQHHKEEHFL VMVVESSLAK TEEESKNGET TDTTLMKFAE TQRNKLETKM 840
841 NDLTDQIFRI LWIFHKIQPM CILVVPRDTL PRRYCSLELA NSTVEKKFLN NDLSAQFVKF 900
901 QFDNVILDFL PHSAYYNESI LSEHLSKLRK MALQEEYAMI EPAYRNGGPV KPKLALQCSG 960
961 VDYRDESVDT RSHTKLTDFK SILEILEWRI SNYGNETAFS DGTNTNLVNS SASNDNNVHK1020
1021 KVSWASFGKI VAGFLKKIVG SKIPLKHGDP IIIMCENSVE YVAMIMACLY CNLLVIPLPS1080
1081 VKESVIEEDL KGLVNIIQSY KVKRVFVDAK LHSLLNDNNV VNKCFKKYKS LIPKITVFSK1140
1141 VKTKNALTVS MFKNVLKQKF GAKPGTRIGM TPCVVWVNTE YDVTSNIHVT MTHSSLLNAS1200
1201 KIVKETLQLR NNSPLFSICS HTSGLGFMFS CLLGIYTGAS TCLFSLTDVL TDPKEFLIGL1260
1261 QNLNVKDLYL KLETLYALLD RASSLIEGFK NKKENINSAK NNTSGSLRED VFKGVRNIMI1320
1321 PFPNRPRIYT IENILKRYST ISLSCTQISY VYQHHFNPLI SLRSYLDIPP VDLYLDPFSL1380
1381 REGIIREVNP NDVSAGNYIK IQDSGVVPVC TDVSVVNPET LLPCVDGEFG EIWCCSEANA1440
1441 FDYFVCNSSK NKLYKDPFIT EQFKSKMKSE VNNTLSYLRT GDLGFIKNVS CTNSQGEVVN1500
1501 LNLLFVLGSI HESIEILGLT HFVSDLERTV KDVHSDIGSC LIAKAGGLLV CLIRCKERHN1560
1561 PILGNLTTLI VSELLNKHGV ILDLCTFVRT KGISPKNSSM IMEVWAKNRA SIMQAWFDQK1620
1621 IQIEAQFGIN YGENISIYLL SDYEKDNI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
CMR2 SSA3 |
| View Details | Krogan NJ, et al. (2006) |

|
CMR2 MTC5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
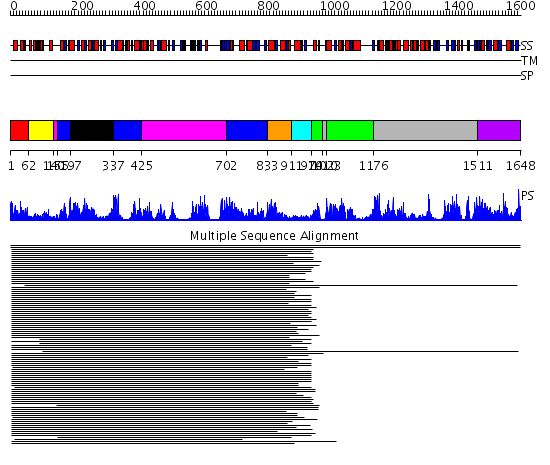
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..61] | 2.20399 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [62..139] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [140..154] [425..701] |
131.0 | Phenylalanine activating domain of gramicidin synthetase 1 | |
| 4 | View Details | [155..196] [337..424] [702..832] |
131.0 | Phenylalanine activating domain of gramicidin synthetase 1 | |
| 5 | View Details | [197..336] | 131.0 | Phenylalanine activating domain of gramicidin synthetase 1 | |
| 6 | View Details | [833..910] | 13.045757 | Peptidyl carrier protein (PCP), thioester domain | |
| 7 | View Details | [911..973] | N/A | Confident ab initio structure predictions are available. | |
| 8 | View Details | [974..1009] [1023..1175] |
79.09691 | Luciferase | |
| 9 | View Details | [1010..1022] [1176..1510] |
79.09691 | Luciferase | |
| 10 | View Details | [1511..1648] | 79.09691 | Luciferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 | No functions predicted. | ||||||||||||
| 2 |
|
||||||||||||
| 3 |
|
||||||||||||
| 4 | No functions predicted. | ||||||||||||
| 5 | No functions predicted. | ||||||||||||
| 6 | No functions predicted. | ||||||||||||
| 7 | No functions predicted. | ||||||||||||
| 8 | No functions predicted. | ||||||||||||
| 9 | No functions predicted. | ||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.39 |
Source: Reynolds et al. (2008)