













| General Information: |
|
| Name(s) found: |
MIG1 /
YGL035C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 504 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA nuclear envelope lumen [IDA |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter by glucose
[IDA |
| Molecular Function: |
sequence-specific DNA binding
[IDA specific transcriptional repressor activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQSPYPMTQV SNVDDGSLLK ESKSKSKVAA KSEAPRPHAC PICHRAFHRL EHQTRHMRIH 60
61 TGEKPHACDF PGCVKRFSRS DELTRHRRIH TNSHPRGKRG RKKKVVGSPI NSASSSATSI 120
121 PDLNTANFSP PLPQQHLSPL IPIAIAPKEN SSRSSTRKGR KTKFEIGESG GNDPYMVSSP 180
181 KTMAKIPVSV KPPPSLALNN MNYQTSSAST ALSSLSNSHS GSRLKLNALS SLQMMTPIAS 240
241 SAPRTVFIDG PEQKQLQQQQ NSLSPRYSNT VILPRPRSLT DFQGLNNANP NNNGSLRAQT 300
301 QSSVQLKRPS SVLSLNDLLV GQRNTNESDS DFTTGGEDEE DGLKDPSNSS IDNLEQDYLQ 360
361 EQSRKKSKTS TPTTMLSRST SGTNLHTLGY VMNQNHLHFS SSSPDFQKEL NNRLLNVQQQ 420
421 QQEQHTLLQS QNTSNQSQNQ NQNQMMASSS SLSTTPLLLS PRVNMINTAI STQQTPISQS 480
481 DSQVQELETL PPIRSLPLPF PHMD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
APP1 CST6 HMO1 MCM10 MIG1 SSA3 YRM1 |
| View Details | Ho Y, et al. (2002) |
|
MIG1 MSS116 NOP12 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 2 | Lockshon D, et al. (2006) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
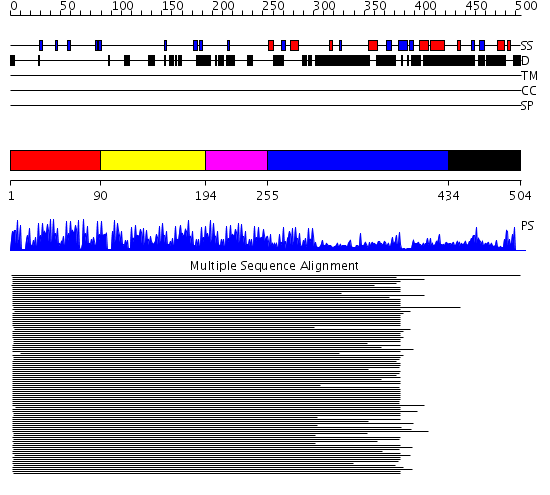
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | 134.70927 | No description for 1lu6A_ was found. | |
| 2 | View Details | [90..193] | 134.70927 | No description for 1lu6A_ was found. | |
| 3 | View Details | [194..254] | 31.033996 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [255..433] | 1.778997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [434..504] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)