













| General Information: |
|
| Name(s) found: |
HCS1 /
YKL017C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 683 amino acids |
Gene Ontology: |
|
| Cellular Component: |
alpha DNA polymerase:primase complex
[IDA |
| Biological Process: |
lagging strand elongation
[IC |
| Molecular Function: |
ATP-dependent 5'-3' DNA helicase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKELASKFL SSIKHEREQD IQTTSRLLTT LSIQQLVQNG LAINNIHLEN IRSGLIGKLY 60
61 MELGPNLAVN DKIQRGDIKV GDIVLVRPAK TKVNTKTKPK VKKVSEDSNG EQAECSGVVY 120
121 KMSDTQITIA LEESQDVIAT TFYSYSKLYI LKTTNVVTYN RMESTMRKLS EISSPIQDKI 180
181 IQYLVNERPF IPNTNSFQNI KSFLNPNLND SQKTAINFAI NNDLTIIHGP PGTGKTFTLI 240
241 ELIQQLLIKN PEERILICGP SNISVDTILE RLTPLVPNNL LLRIGHPARL LDSNKRHSLD 300
301 ILSKKNTIVK DISQEIDKLI QENKKLKNYK QRKENWNEIK LLRKDLKKRE FKTIKDLIIQ 360
361 SRIVVTTLHG SSSRELCSLY RDDPNFQLFD TLIIDEVSQA MEPQCWIPLI AHQNQFHKLV 420
421 LAGDNKQLPP TIKTEDDKNV IHNLETTLFD RIIKIFPKRD MVKFLNVQYR MNQKIMEFPS 480
481 HSMYNGKLLA DATVANRLLI DLPTVDATPS EDDDDTKIPL IWYDTQGDEF QETADEATIL 540
541 GSKYNEGEIA IVKEHIENLR SFNVPENSIG VISPYNAQVS HLKKLIHDEL KLTDIEISTV 600
601 DGFQGREKDV IILSLVRSNE KFEVGFLKEE RRLNVAMTRP RRQLVVVGNI EVLQRCGNKY 660
661 LKSWSEWCEE NADVRYPNID DYL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
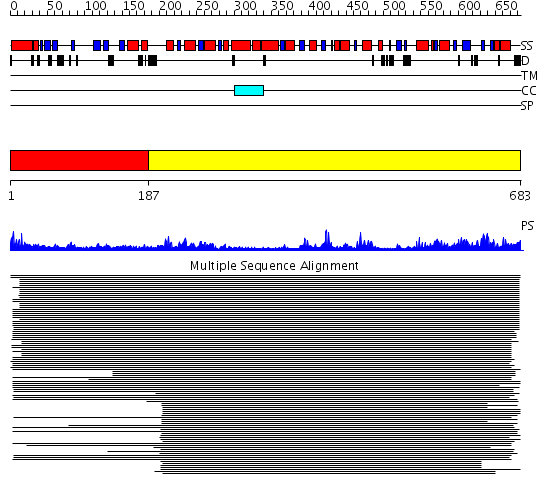
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..186] | 1.212988 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [187..683] | 70.154902 | DEXX box DNA helicase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)