













| General Information: |
|
| Name(s) found: |
PEP1 /
YBL017C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1579 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Golgi apparatus
[TAS |
| Biological Process: |
vacuolar transport
[IMP protein targeting to vacuole [IMP |
| Molecular Function: |
signal sequence binding
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MILLHFVYSL WALLLIPLTN AEEFTPKVTK TIAQDSFDIL SFDDSNTLIR KQDTSVTISF 60
61 DDGETWEKVE GIEGEITWIY IDPFNRHDRA VATAMNGSYL YITNDQGKSW ERITLPDSGE 120
121 SISPRECYIE THPLNKNYFL AKCNYCEKTE VNNDNEENSG DEEGQFEIFN ITRCTDKVFA 180
181 SNDGGKSFSE IKSSLERNEN SPISISDCGF AKTSKDSDLE SSDTSIICLF QNMQLIMDEF 240
241 SSPYTESKLV LTTDWGKSLK EFDQFKDKVV NGYRILKSHM VVLTQGDRYN DMSSMDVWVS 300
301 NDLSNFKMAY MPTQLRHSMQ GEIYEDAMGR IILPMSRERS DQEEDKGIVS EILISDSQGL 360
361 KFSPIPWTAN EVFGYINFYQ PTYLKGTMIA SLYPLSRRRN RKGKAKGVKS KGVTKISVDN 420
421 GLTWTMLKVV DPDNADSFDC DITDFENCSL QNMFYTREGS TPTAGILMTT GIVGDGSVFD 480
481 WGDQRTFISR DGGLTWKLAF DFPCLYAVGD YGNVIVAIPY NADEDDDPQS EFYYSLDQGK 540
541 TWTEYQLETT IYPNEVMNTT PDGSGAKFIL NGFTLAHMDG TTNFIYAIDF STAFNDKTCE 600
601 ENDFEDWNLA EGKCVNGVKY KIRRRKQDAQ CLVKKVFEDL QLFETACDKC TEADYECAFE 660
661 FVRDATGKCV PDYNLIVLSD VCDKTKKKTV PVKPLQLVKG DKCKKPMTVK SVDISCEGVP 720
721 KKGTNDKEIV VTENKFDFKI QFYQYFDTVT DESLLMINSR GEAYISHDGG QTIKRFDSNG 780
781 ETIIEVVFNP YYNSSAYLFG SKGSIFSTHD RGYSFMTAKL PEARQLGMPL DFNAKAQDTF 840
841 IYYGGKNCES ILSPECHAVA YLTNDGGETF TEMLDNAIHC EFAGSLFKYP SNEDMVMCQV 900
901 KEKSSQTRSL VSSTDFFQDD KNTVFENIIG YLSTGGYIIV AVPHENNELR AYVTIDGTEF 960
961 AEAKFPYDED VGKQEAFTIL ESEKGSIFLH LATNLVPGRD FGNLLKSNSN GTSFVTLEHA1020
1021 VNRNTFGYVD FEKIQGLEGI ILTNIVSNSD KVAENKEDKQ LKTKITFNEG SDWNFLKPPK1080
1081 RDSEGKKFSC SSKSLDECSL HLHGYTERKD IRDTYSSGSA LGMMFGVGNV GPNLLPYKEC1140
1141 STFFTTDGGE TWAEVKKTPH QWEYGDHGGI LVLVPENSET DSISYSTDFG KTWKDYKFCA1200
1201 DKVLVKDITT VPRDSALRFL LFGEAADIGG SSFRTYTIDF RNIFERQCDF DITGKESADY1260
1261 KYSPLGSKSN CLFGHQTEFL RKTDENCFIG NIPLSEFSRN IKNCSCTRQD FECDYNFYKA1320
1321 NDGTCKLVKG LSPANAADVC KKEPDLIEYF ESSGYRKIPL STCEGGLKLD APSSPHACPG1380
1381 KEKEFKEKYS VSAGPFAFIF ISILLIIFFA AWFVYDRGIR RNGGFARFGE IRLGDDGLIE1440
1441 NNNTDRVVNN IVKSGFYVFS NIGSLLQHTK TNIAHAISKI RGRFGNRTGP SYSSLIHDQF1500
1501 LDEADDLLAG HDEDANDLSS FMDQGSNFEI EEDDVPTLEE EHTSYTDQPT TTDVPDTLPE1560
1561 GNEENIDRPD STAPSNENQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
CSI1 CSN9 ERV46 FPR2 HUL5 IQG1 MOT1 MPT5 NAS6 NMD3 NPT1 PCI8 PEP1 PST1 PUS9 RPN1 RPN10 RPN11 RPN12 RPN13 RPN14 RPN2 RPN5 RPN6 RPN7 RPN8 RPN9 RPT1 RPT2 RPT3 RPT4 RPT5 RPT6 RRI1 RRI2 SEC18 SEM1 SKP1 SPG5 SPO77 SQT1 SWA2 TCB2 TIP41 TOD6 TOR2 TSR4 UBP6 YEN1 YKL027W YMR258C YNL022C YPR003C YPT52 |
| View Details | Gavin AC, et al. (2006) |
|
DIG1 NPL3 PEP1 |
| View Details | Gavin AC, et al. (2002) |
|
DIG1 NPL3 PEP1 |
| View Details | Ho Y, et al. (2002) |
|
AAC3 BZZ1 GAL2 HXT6 HXT7 LAS17 MYO4 PEP1 PHO84 RPN1 RPN12 RVS167 SLA1 SQT1 VMA6 VRP1 YHM2 YNR065C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
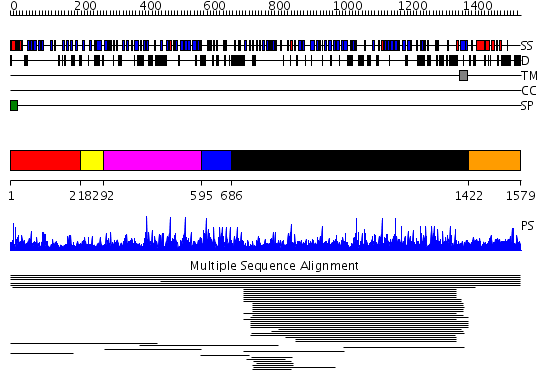
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..217] | 1.005944 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [218..291] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [292..594] | 1.007897 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [595..685] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [686..1421] | 7.020001 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [1422..1579] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||
| 6 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|