













| General Information: |
|
| Name(s) found: |
MKS1 /
YNL076W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 584 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IC |
| Biological Process: |
regulation of nitrogen utilization
[IMP mitochondria-nucleus signaling pathway [IPI |
| Molecular Function: |
transcription repressor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSREAFDVPN IGTNKFLKVT PNLFTPERLN LFDDVELYLT LIKASKCVEQ GERLHNISWR 60
61 ILNKAVLKEH NINRSKKRDG VKNIYYVLNP NNKQPIKPKQ AAVKQPPLQK ANLPPTTAKQ 120
121 NVLTRPMTSP AIAQGAHDRS LDNPNSTNND VKNDVAPNRQ FSKSTTSGLF SNFADKYQKM 180
181 KNVNHVANKE EPQTIITGFD TSTVITKKPL QSRRSRSPFQ HIGDMNMNCI DNETSKSTSP 240
241 TLENMGSRKS SFPQKESLFG RPRSYKNDQN GQLSLSKTSS RKGKNKIFFS SEDEDSDWDS 300
301 VSNDSEFYAD EDDEEYDDYN EEEADQYYRR QWDKLLFAKN QQNLDSTKSS VSSANTINSN 360
361 TSHDPVRKSL LSGLFLSEAN SNSNNHNTAH SEYASKHVSP TPQSSHSNIG PQPQQNPPSA 420
421 NGIKQQKPSL KTSNVTALAS LSPPQPSNNE RLSMDIQKDF KTDNESNHLY ESNAPLTAQT 480
481 ILPTALSTHM FLPNNIHQQR MAIATGSNTR HRFSRRQSMD IPSKNRNTGF LKTRMEISEE 540
541 EKMVRTISRL DNTSIANSNG NGNDDTSNQR TEALGRKTSN GGRI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
MKS1 RTG2 YNL208W |
| View Details | Gavin AC, et al. (2006) |
|
GLN1 MCM2 MKS1 RTG2 TSC10 |
| View Details | Gavin AC, et al. (2002) |
|
CTR3 DAL3 GLN1 GTO3 IML1 MCM2 MKS1 REG1 TIF1, TIF2 TPS1 TPS2 TPS3 TSC10 TSL1 YIL177C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
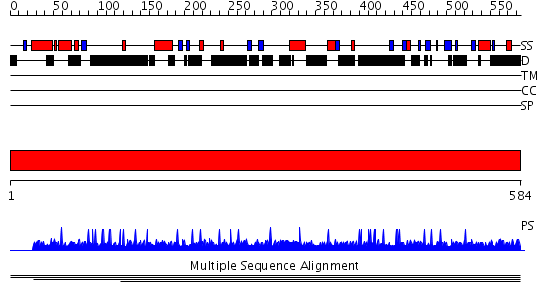
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..584] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)