













| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRIRSLLNN KRAFSSTVRT LTINKSHDVV IIGGGPAGYV AAIKAAQLGF NTACVEKRGK 60
61 LGGTCLNVGC IPSKALLNNS HLFHQMHTEA QKRGIDVNGD IKINVANFQK AKDDAVKQLT 120
121 GGIELLFKKN KVTYYKGNGS FEDETKIRVT PVDGLEGTVK EDHILDVKNI IVATGSEVTP 180
181 FPGIEIDEEK IVSSTGALSL KEIPKRLTII GGGIIGLEMG SVYSRLGSKV TVVEFQPQIG 240
241 ASMDGEVAKA TQKFLKKQGL DFKLSTKVIS AKRNDDKNVV EIVVEDTKTN KQENLEAEVL 300
301 LVAVGRRPYI AGLGAEKIGL EVDKRGRLVI DDQFNSKFPH IKVVGDVTFG PMLAHKAEEE 360
361 GIAAVEMLKT GHGHVNYNNI PSVMYSHPEV AWVGKTEEQL KEAGIDYKIG KFPFAANSRA 420
421 KTNQDTEGFV KILIDSKTER ILGAHIIGPN AGEMIAEAGL ALEYGASAED VARVCHAHPT 480
481 LSEAFKEANM AAYDKAIHC |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
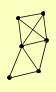
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
KGD1 KGD2 LAT1 LPD1 PDB1 YMR31 |
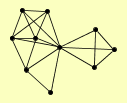
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
GCV1 GCV2 GCV3 KGD1 KGD2 LAT1 LPD1 PDA1 PDB1 PDX1 |
| View Details | Krogan NJ, et al. (2006) |

|
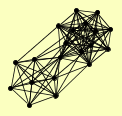
LPD1 YMR31 |
| View Details | Gavin AC, et al. (2002) |
|
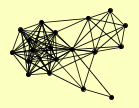
APM1 CLU1 CYM1 DOA4 DUR1,2 HSP42 KAP123 KGD1 KGD2 LAT1 LEA1 LPD1 PDA1 PDB1 PDX1 PRP46 RPN11 SAM1 TCM62 YMR31 |
| View Details | Ho Y, et al. (2002) |
|
AFG3 ARC1 ARG1 CDC10 CDC11 CDC12 CDC3 CDC33 COR1 ENP1 KGD2 KRS1 LPD1 PMI40 RVB2 RVS161 RVS167 SES1 TRP5 |
| View Details | Qiu et al. (2008) |
|
ALD4 ATP1 GCV1 GCV2 GCV3 ILV5 KGD1 LAT1 LPD1 MDH1 PDA1 PDB1 PDX1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | Boi 2 gst control | McCusker D, et al (2007) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
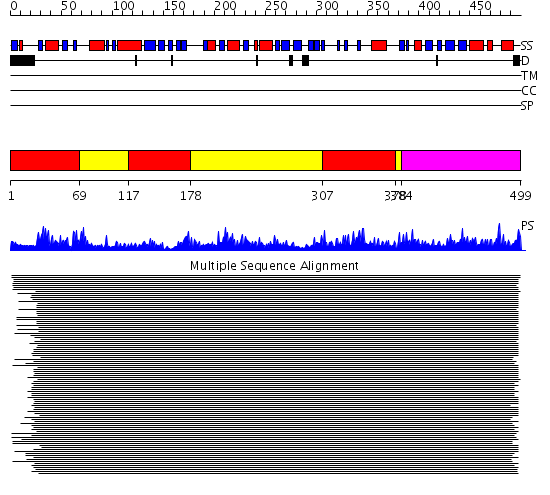
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..68] [117..177] [307..377] |
10160.0 | Dihydrolipoamide dehydrogenase | |
| 2 | View Details | [69..116] [178..306] [378..383] |
10160.0 | Dihydrolipoamide dehydrogenase | |
| 3 | View Details | [384..499] | 10160.0 | Dihydrolipoamide dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)