













| General Information: |
|
| Name(s) found: |
CYM1 /
YDR430C
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 989 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial intermembrane space
[IDA mitochondrion [IDA |
| Biological Process: |
proteolysis
[IDA |
| Molecular Function: |
metalloendopeptidase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRFQRFASS YAQAQAVRKY PVGGIFHGYE VRRILPVPEL RLTAVDLVHS QTGAEHLHID 60
61 RDDKNNVFSI AFKTNPPDST GVPHILEHTT LCGSVKYPVR DPFFKMLNKS LANFMNAMTG 120
121 PDYTFFPFST TNPQDFANLR GVYLDSTLNP LLKQEDFDQE GWRLEHKNIT DPESNIVFKG 180
181 VVYNEMKGQI SNANYYFWSK FQQSIYPSLN NSGGDPMKIT DLRYGDLLDF HHKNYHPSNA 240
241 KTFTYGNLPL VDTLKQLNEQ FSGYGKRARK DKLLMPIDLK KDIDVKLLGQ IDTMLPPEKQ 300
301 TKASMTWICG APQDTYDTFL LKVLGNLLMD GHSSVMYQKL IESGIGLEFS VNSGVEPTTA 360
361 VNLLTVGIQG VSDIEIFKDT VNNIFQNLLE TEHPFDRKRI DAIIEQLELS KKDQKADFGL 420
421 QLLYSILPGW TNKIDPFESL LFEDVLQRFR GDLETKGDTL FQDLIRKYIV HKPCFTFSIQ 480
481 GSEEFSKSLD DEEQTRLREK ITALDEQDKK NIFKRGILLQ EKQNEKEDLS CLPTLQIKDI 540
541 PRAGDKYSIE QKNNTMSRIT DTNGITYVRG KRLLNDIIPF ELFPYLPLFA ESLTNLGTTT 600
601 ESFSEIEDQI KLHTGGISTH VEVTSDPNTT EPRLIFGFDG WSLNSKTDHI FEFWSKILLE 660
661 TDFHKNSDKL KVLIRLLASS NTSSVADAGH AFARGYSAAH YRSSGAINET LNGIEQLQFI 720
721 NRLHSLLDNE ETFQREVVDK LTELQKYIVD TNNMNFFITS DSDVQAKTVE SQISKFMERL 780
781 PHGSCLPNGP KTSDYPLIGS KCKHTLIKFP FQVHYTSQAL LGVPYTHKDG SALQVMSNML 840
841 TFKHLHREVR EKGGAYGGGA SYSALAGIFS FYSYRDPQPL KSLETFKNSG RYILNDAKWG 900
901 VTDLDEAKLT IFQQVDAPKS PKGEGVTYFM SGVTDDMKQA RREQLLDVSL LDVHRVAEKY 960
961 LLNKEGVSTV IGPGIEGKTV SPNWEVKEL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |

|
CYM1 MSS51 |
| View Details | Gavin AC, et al. (2002) |
|
APM1 CYM1 DOA4 DUR1,2 HSP42 LAT1 LPD1 PDA1 PDB1 PDX1 PRP46 RPN11 TCM62 |
| View Details | Qiu et al. (2008) |
|
CYM1 PRD1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
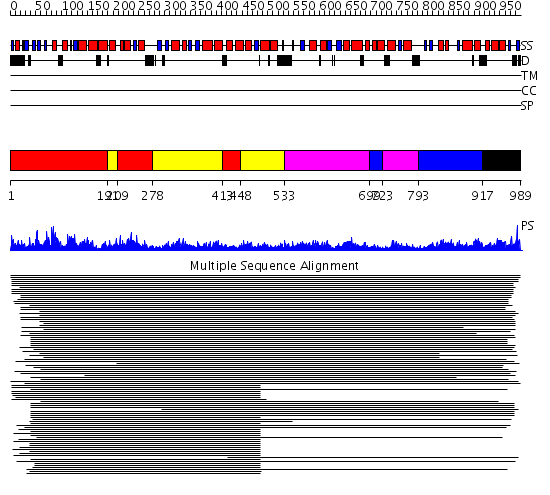
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] [209..277] [413..447] |
91.69897 | Cytochrome bc1 core subunit 1 | |
| 2 | View Details | [191..208] [278..412] [448..532] |
91.69897 | Cytochrome bc1 core subunit 1 | |
| 3 | View Details | [533..698] [723..792] |
19.0 | Cytochrome bc1 core subunit 2 | |
| 4 | View Details | [699..722] [793..916] |
19.0 | Cytochrome bc1 core subunit 2 | |
| 5 | View Details | [917..989] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)