













| General Information: |
|
| Name(s) found: |
VAC8 /
YEL013W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 578 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type vacuole
[IDA |
| Biological Process: |
vacuole inheritance
[IMP vacuole fusion, non-autophagic [IDA protein targeting to vacuole [IMP microautophagy [IMP |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSCCSCLKD SSDEASVSPI ADNEREAVTL LLGYLEDKDQ LDFYSGGPLK ALTTLVYSDN 60
61 LNLQRSAALA FAEITEKYVR QVSREVLEPI LILLQSQDPQ IQVAACAALG NLAVNNENKL 120
121 LIVEMGGLEP LINQMMGDNV EVQCNAVGCI TNLATRDDNK HKIATSGALI PLTKLAKSKH 180
181 IRVQRNATGA LLNMTHSEEN RKELVNAGAV PVLVSLLSST DPDVQYYCTT ALSNIAVDEA 240
241 NRKKLAQTEP RLVSKLVSLM DSPSSRVKCQ ATLALRNLAS DTSYQLEIVR AGGLPHLVKL 300
301 IQSDSIPLVL ASVACIRNIS IHPLNEGLIV DAGFLKPLVR LLDYKDSEEI QCHAVSTLRN 360
361 LAASSEKNRK EFFESGAVEK CKELALDSPV SVQSEISACF AILALADVSK LDLLEANILD 420
421 ALIPMTFSQN QEVSGNAAAA LANLCSRVNN YTKIIEAWDR PNEGIRGFLI RFLKSDYATF 480
481 EHIALWTILQ LLESHNDKVE DLVKNDDDII NGVRKMADVT FERLQRSGID VKNPGSNNNP 540
541 SSNDNNSNNN DTGSEHQPVE DASLELYNIT QQILQFLH |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
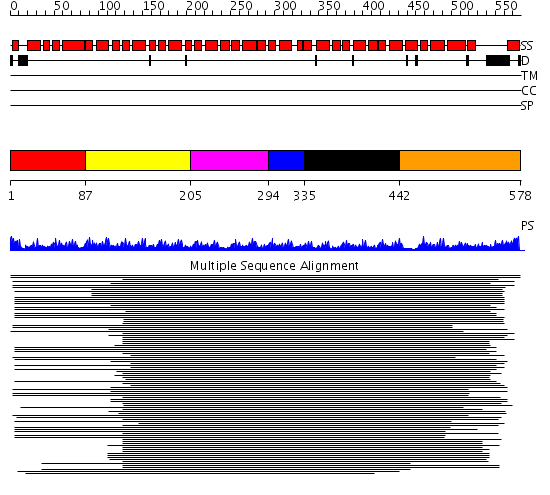
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | 123.0 | beta-Catenin | |
| 2 | View Details | [87..204] | 123.0 | beta-Catenin | |
| 3 | View Details | [205..293] | 123.0 | beta-Catenin | |
| 4 | View Details | [294..334] | 123.0 | beta-Catenin | |
| 5 | View Details | [335..441] | 123.0 | beta-Catenin | |
| 6 | View Details | [442..578] | 194.28043 | Karyopherin alpha |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)