













| General Information: |
|
| Name(s) found: |
CAT8 /
YMR280C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1433 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
positive regulation of transcription from RNA polymerase II promoter
[IDA positive regulation of gluconeogenesis [IMP |
| Molecular Function: |
specific RNA polymerase II transcription factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANNNSDRQG LEPRVIRTLG SQALSGPSIS NRTSSSEANP HFSKNVKEAM IKTASPTPLS 60
61 TPIYRIAQAC DRCRSKKTRC DGKRPQCSQC AAVGFECRIS DKLLRKAYPK GYTESLEERV 120
121 RELEAENKRL LALCDIKEQQ ISLVSQSRPQ TSTDNTINGN FKHDLKDAPL NLSSTNIYLL 180
181 NQTVNKQLQN GKMDGDNSGS AMSPLGAPPP PPHKDHLCDG VSCTNHLHVK PTSTSLNDPT 240
241 AISFEQDEAP GLPAVKALKS MTTHQRSTQL ATLVSLSIPR STEEILFIPQ LLTRIRQIFG 300
301 FNSKQCLYTV SLLSSLKNRL PAPRLLAPST STKLKEKDED KKLDDDSAFV KRFQSTNLSE 360
361 FVDLKKFLIS LKFNINSFSK QSEKPANDQD DELLSLTEIK ELLHLFFKFW SNQVPILNND 420
421 HFLIYFNNFV EVVKHLSTEN LETNNTTKST VTTNHEIFAL KLLMMLQMGL LVKIKMEKIK 480
481 YTVPKNPKAK YARLMAYYHQ LSLIIPKNPY FLNMSTTSLP SLQLLSLASF YYLNVGDISA 540
541 IYGVRGRIVS MAQQLRLHRC PSAVLSVHSN PVLQKFEQSE RRLLFWAIYY VDVFASLQLG 600
601 VPRLLKDFDI ECALPISDVE YKDQLSMENE KADKKAKKIQ LQGQVSSFSL QIIRFAKILG 660
661 NILDSIFKRG MMDERITSEV ALVHENALDN WRNQLPEMYY FQITVNGTVN LDEIRATNQR 720
721 NTETKFDKKD IILFEKKILL LFYFLAKSMI HLPVIATKPL PKNVDNATKK KQSMFNNDSK 780
781 GATNQDHMIL DVDMTSPAIR TSSSYIILQQ ATNATLTIFQ AINSMYLPLP LNVSRTLIRF 840
841 SLLCARGSLE YTKGGALFLD NKNLLLDTIK DIENDRLLDL PGIASWHTLK LFDMSINLLL 900
901 KAPNVKVERL DKFLEKKLNY YNRLMGLPPA TTTSLKPLFG SQSKNSLENR QRTPNVKREN 960
961 PEHEYLYGND SNNNNNSEAG HSPMTNTTNG NKRLKYEKDA KRNAKDGGIS KGENAHNFQN1020
1021 DTKKNMSTSN LFPFSFSNTD LTALFTHPEG PNCTNTNNGN VDVCNRASTD ATDANIENLS1080
1081 FLNMAPFLQT GNSNIGQNTI ENKPMHMDAI FSLPSNLDLM KDNMDSKPEQ LEPVIKQNPE1140
1141 NSKNNQFHQK GKSTNMEKNN LSFNNKSNYS LTKLMRLLNN DNSFSNISIN NFLYQNDQNS1200
1201 ASADPGTNKK AVTNAGANFK PPSTGSNTSQ GSILGSTKHG MDNCDFNDLG NFNNFMTNVN1260
1261 YSGVDYDYIV DASLGLAPLL VDTPDISNTN TTSTTSNRSK NSIILDTTFN DDLDRSRMNA1320
1321 REVLNPTDSI LSQGMVSSVS TRNTSNQRSL SSGNDSKGDS SSQENSKSAT GNQLDTPSTL1380
1381 FQMRRTSSGP SASHRGPRRP QKNRYNTDRS KSSGGGSSNT DNVSDLFQWQ NAK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | sample bir 1 from feb 2005 | Sandall S, et all (2006) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
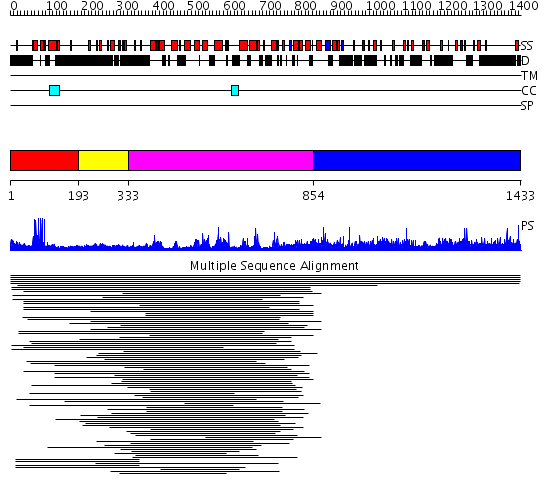
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..192] | 81.144602 | Gal4; CD2-Gal4 | |
| 2 | View Details | [193..332] | 1.084993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [333..853] | 8.106973 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [854..1433] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [513..764] | 2.967994 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [765..824] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [825..1044] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1045..1206] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1207..1433] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)