













| General Information: |
|
| Name(s) found: |
YKL050C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 922 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLISALQTT DVESVQTSPE QITERKAVRV STLQESLHSS EMHRAAPETP RSISNSVHKL 60
61 KTIYSTYQQS GQPLSKEAIF RAKQKYGILN TPANYKTLGL GDSKSESVDL AARLASKRTK 120
121 VSPDDCVETA IEQKARGEAF KVTFSKIPLT PPEDVPITVN LGLKGRRDFL TRLAAQKALA 180
181 FSPSLDNSMK GTSDSSSVKK KRFSGAPIGN EFDANLVNPQ HPAGFKSLDL SKVLDGAERR 240
241 AISRVNDRLY PQKVNFKNGL QSSDQSGVSK ANKEVFKKGT LEKLEHSAEQ FLESHAGNER 300
301 QRLSDQQYMC AKGAADAVKD LDPKTLEDPD FAAREAQKKL YIKQVASPVV LNEAQKLANR 360
361 KLQDIDSRDT YMLLFGNQAY NKLAVNIALQ HYSVKQEEKK KIYLGGGLWM TPEEVNAVAK 420
421 KLISPVVNEI DERASRQRDV DKDIERRSRV LDQEYEDGNS MERAKEQNDG QLLLAMASKQ 480
481 QQEKEAKKAE EGQRYDQFVQ KMNIKLQQKE KELENARENR ENLRNELQER LSKNLSGEND 540
541 ELNDWNDACE RDLKNSSIEH YYAVRSHFDN LGNSERGYDE LLEERSKIQV EIERLVASIA 600
601 EHKTAIHGFG ETADAGGAIP AVQKQKIPTR KDLLDATVND PLVISAEMAK EEAEMATEEC 660
661 MLKELQVDEM IIIRNIMLRE CEKKLEEEKE TAKRSRRGTE ESKNNSNFSR DVIMSTPDNN 720
721 EKVTPIGKSA SPKDVVKSRF LSTYNTGKDI DSSASARSIT GVSGVLDDGP KTPTSNKENE 780
781 LIDDEVKSYK VHQAVDGTGE DSIANKRDKS SRPAANSGGS ITIEQFLFNK NADKQGLSKT 840
841 ESVTMKREPV VDQMDSKKGH DFTHCNDNGR RSFSGFSQGS IENDYSNEVT DDQDDQEGSE 900
901 IRVRDSNDSN TSPKESFFKE VI |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
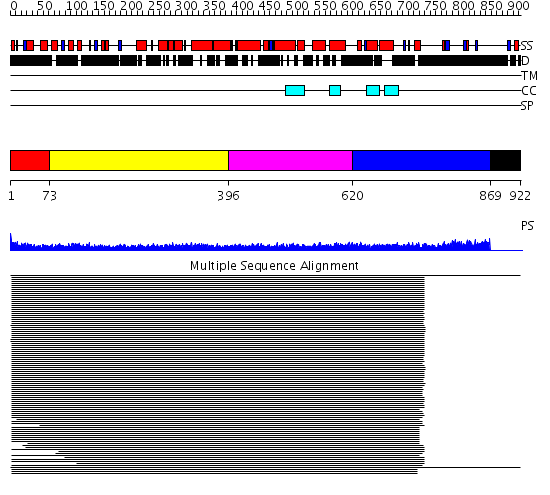
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..72] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [73..395] | 13.154902 | Heavy meromyosin subfragment | |
| 3 | View Details | [396..619] | 8.045757 | Tropomyosin | |
| 4 | View Details | [620..868] | 4.011996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [869..922] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)