













| General Information: |
|
| Name(s) found: |
GID8 /
YMR135C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 455 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
negative regulation of gluconeogenesis
[IMP traversing start control point of mitotic cell cycle [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTISTLSNET TKSGSCSGQG KNGGKDFTYG KKCFTKEEWK EQVAKYSAMG ELYANKTIHY 60
61 PLKIQPNSSG GSQDEGFATI QTTPIEPTLP RLLLNYFVSM AYEDSSIRMA KELGFIRNNK 120
121 DIAVFNDLYK IKERFHIKHL IKLGRINEAM EEINSIFGLE VLEETFNATG SYTGRTDRQQ 180
181 QQQQQQFDID GDLHFKLLLL NLIEMIRSHH QQENITKDSN DFILNLIQYS QNKLAIKASS 240
241 SVKKMQELEL AMTLLLFPLS DSADSGSIKL PKSLQNLYSI SLRSKIADLV NEKLLKFIHP 300
301 RIQFEISNNN SKFPDLLNSD KKIITQNFTV YNNNLVNGSN GTKITHISSD QPINEKMSSN 360
361 EVTAAANSVW LNQRDGNVGT GSAATTFHNL ENKNYWNQTS ELLSSSNGKE KGLEFNNYYS 420
421 SEFPYEPRLT QIMKLWCWCE NQLHHNQIGV PRVEN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
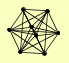
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
FYV10 GID7 GID8 RMD5 VID24 VID28 VID30 YDL176W |
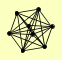
| View Details | Krogan NJ, et al. (2006) |
|
FYV10 GID7 GID8 RMD5 VID24 VID28 VID30 YDL176W |
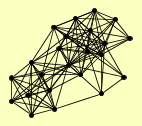
| View Details | Ho Y, et al. (2002) |
|
ASF1 CCT2 CCT3 CTF19 FYV10 GID7 GID8 HTA2 HXT7 IPP1 MDH1 MOH1 PAA1 RAD53 RMD5 SHS1 SMC3 TFP1 UME1 UTP22 VID24 VID28 VID30 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
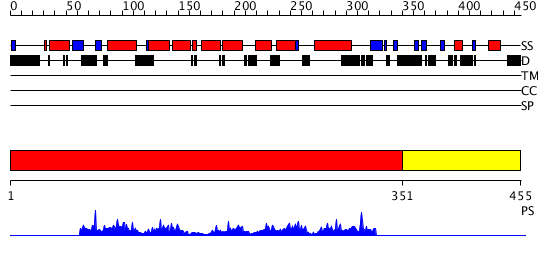
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..350] | 13.025968 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [351..455] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)