













| General Information: |
|
| Name(s) found: |
VID30 /
YGL227W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 958 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
proteasomal ubiquitin-dependent protein catabolic process
[IMP negative regulation of gluconeogenesis [IMP regulation of nitrogen utilization [IMP vacuolar protein catabolic process [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEYMDDVDR EFINCLFPSY LLQQPVAYDL WILYLQHRKL FHKLKNTNLI NADENPTGVG 60
61 MGRTKLTALT RKEIWSKLMN LGVLGTISFE AVNDDYLIQV YKYFYPDVND FTLRFGVKDS 120
121 NKNSVRVMKA SSDMRKNAQE LLEPVLSERE MALNSNTSLE NDRNDDDDDD DDDDDDDDDD 180
181 DDDDDESDLE SLEGEVDTDT DDNNEGDGSD NHEEGGEEGS RGADADVSSA QQRAERVADP 240
241 WIYQRSRSAI NIETESRNLW DTSDKNSGLQ YYPPDQSPSS SFSSPRVSSG NDKNDNEATN 300
301 VLSNSGSKKK NSMIPDIYKI LGYFLPSRWQ AQPNNSLQLS QDGITHLQPN PDYHSYMTYE 360
361 RSSASSASTR NRLRTSFENS GKVDFAVTWA NKSLPDNKLT IFYYEIKVLS VTSTESAENS 420
421 NIVIGYKLVE NELMEATTKK SVSRSSVAGS SSSLGGSNNM SSNRVPSTSF TMEGTQRRDY 480
481 IYEGGVSAMS LNVDGSINKC QKYGFDLNVF GYCGFDGLIT NSTEQSKEYA KPFGRDDVIG 540
541 CGINFIDGSI FFTKNGIHLG NAFTDLNDLE FVPYVALRPG NSIKTNFGLN EDFVFDIIGY 600
601 QDKWKSLAYE HICRGRQMDV SIEEFDSDES EEDETENGPE ENKSTNVNED LMDIDQEDGA 660
661 AGNKDTKKLN DEKDNNLKFL LGEDNRFIDG KLVRPDVNNI NNLSVDDGSL PNTLNVMIND 720
721 YLIHEGLVDV AKGFLKDLQK DAVNVNGQHS ESKDVIRHNE RQIMKEERMV KIRQELRYLI 780
781 NKGQISKCIN YIDNEIPDLL KNNLELVFEL KLANYLVMIK KSSSKDDDEI ENLILKGQEL 840
841 SNEFIYDTKI PQSLRDRFSG QLSNVSALLA YSNPLVEAPK EISGYLSDEY LQERLFQVSN 900
901 NTILTFLHKD SECALENVIS NTRAMLSTLL EYNAFGSTNS SDPRYYKAIN FDEDVLNL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
FYV10 GID7 GID8 RMD5 VID24 VID28 VID30 YDL176W |
| View Details | Krogan NJ, et al. (2006) |
|
FYV10 GID7 GID8 RMD5 VID24 VID28 VID30 YDL176W |
| View Details | Ho Y, et al. (2002) |
|
CCT2 CCT3 CTF19 FYV10 GID7 GID8 HXT7 MOH1 RMD5 TFP1 UME1 VID28 VID30 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
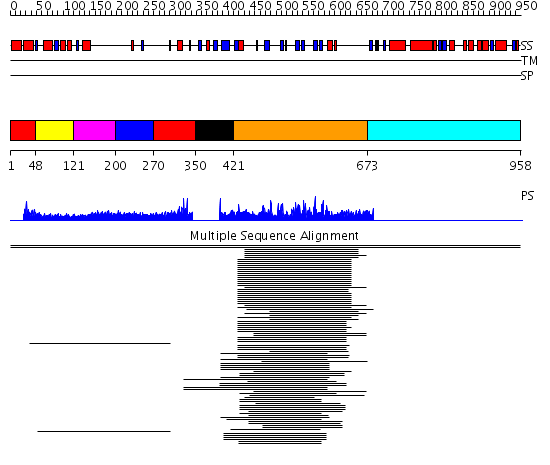
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..47] [270..349] |
10.3 | No description for 1l8mA was found. | |
| 2 | View Details | [48..120] | 10.3 | No description for 1l8mA was found. | |
| 3 | View Details | [121..199] | 10.3 | No description for 1l8mA was found. | |
| 4 | View Details | [200..269] | 10.3 | No description for 1l8mA was found. | |
| 5 | View Details | [350..420] | N/A | Confident ab initio structure predictions are available. | |
| 6 | View Details | [421..672] | 36.103997 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [673..958] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)