













| General Information: |
|
| Name(s) found: |
ADE16 /
YLR028C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 591 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
ascospore formation
[IEP purine nucleotide biosynthetic process [IDA 'de novo' IMP biosynthetic process [IDA aerobic respiration [IEP |
| Molecular Function: |
IMP cyclohydrolase activity
[IDA phosphoribosylaminoimidazolecarboxamide formyltransferase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKYTKTAIL SVYDKTGLLD LAKGLVENNV RILASGGTAN MVREAGFPVD DVSSITHAPE 60
61 MLGGRVKTLH PAVHAGILAR NLEGDEKDLK EQHIDKVDFV VCNLYPFKET VAKIGVTVQE 120
121 AVEEIDIGGV TLLRAAAKNH SRVTILSDPN DYSIFLQDLS KDGEISQDLR NRFALKAFEH 180
181 TADYDAAISD FFRKQYSEGK AQLPLRYGCN PHQRPAQAYI TQQEELPFKV LCGTPGYINL 240
241 LDALNSWPLV KELSASLNLP AAASFKHVSP AGAAVGLPLS DVERQVYFVN DMEDLSPLAC 300
301 AYARARGADR MSSFGDFIAL SNIVDVATAK IISKEVSDGV IAPGYEPEAL NILSKKKNGK 360
361 YCILQIDPNY VPGQMESREV FGVTLQQKRN DAIINQSTFK EIVSKNKALT EQAVIDLTVA 420
421 TLVLKYTQSN SVCYAKNGMV VGLGAGQQSR IHCTRLAGDK TDNWWLRQHP KVLNMKWAKG 480
481 IKRADKSNAI DLFVTGQRIE GPEKVDYESK FEEVPEPFTK EERLEWLSKL NNVSLSSDAF 540
541 FPFPDNVYRA VQSGVKFITA PSGSVMDKVV FQAADSFDIV YVENPIRLFH H |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
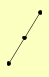
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ADE16 ADE17 GLC7 |
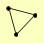
| View Details | Krogan NJ, et al. (2006) |
|
ADE16 ADE17 SLX1 |
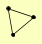
| View Details | Gavin AC, et al. (2006) |
|
ADE16 GLC7 SOL2 |
| View Details | Qiu et al. (2008) |

|
ADE16 ADE17 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
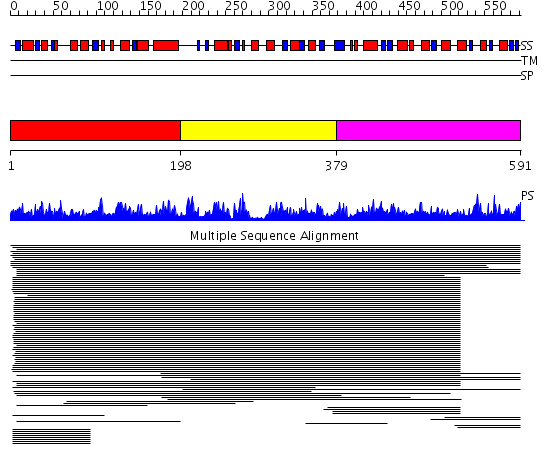
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..197] | 11000.0 | IMP cyclohydrolase domain of bifunctional purine biosynthesis enzyme ATIC; AICAR transformylase domain of bifunctional purine biosynthesis enzyme ATIC | |
| 2 | View Details | [198..378] | 11000.0 | IMP cyclohydrolase domain of bifunctional purine biosynthesis enzyme ATIC; AICAR transformylase domain of bifunctional purine biosynthesis enzyme ATIC | |
| 3 | View Details | [379..591] | 11000.0 | IMP cyclohydrolase domain of bifunctional purine biosynthesis enzyme ATIC; AICAR transformylase domain of bifunctional purine biosynthesis enzyme ATIC |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)