













| General Information: |
|
| Name(s) found: |
SLX1 /
YBR228W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 304 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
DNA-dependent DNA replication
[IGI response to DNA damage stimulus [IGI |
| Molecular Function: |
5'-flap endonuclease activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQKIQQHQF PDFYCCYLLQ SINKRQSFYV GSTPNPVRRL RQHNGKLAVG GAYRTKRDGS 60
61 RPWEMIMIVR GFPSKIAALQ FEHAWQHGYQ THYIAEKDRV VKHKAGGRTL HHKVALMKLL 120
121 LKHEFFQRMN LIVEVFNIKA WEVWKQDKFF IERDRFPINI QINENALEEP KEKTVDVLMD 180
181 HSDENLKVVE AVYTKVIENE RNIFETFEKK LTTGVVRCEI CEKEIDYTSE EQNLKPFVAL 240
241 CNNKDCGCVN HLKCLHRYFL DDEQLMVGRR NLIPRGGKCP KCDMFCDWTT LVKFSTRMKL 300
301 AHGK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
ADE16 ADE17 SLX1 |
| View Details | Ho Y, et al. (2002) |

|
ATP3 AVO1 CCT5 CLU1 DHH1 DPB2 FUN12 GAL7 GPH1 IDH1 ILV2 MET16 MKK2 NOG2 NPA3 PDX1 PHO85 PUF3 RPN12 SIS1 SLX1 SRP54 STI1 TEM1 TFC7 VMA8 YER077C YGR266W YKR051W YKU80 YLR271W YPR003C |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
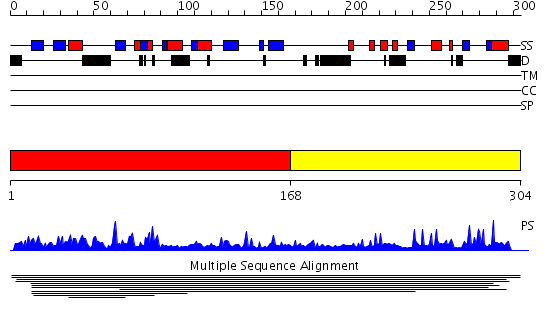
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..167] | 2.00998 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [168..304] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [180..304] | 2.29 | Solution Structure of the Ring-H2 Finger Domain of Mouse Deltex Protein 2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)