













| General Information: |
|
| Name(s) found: |
GAL7 /
YBR018C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 366 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
galactose catabolic process
[IDA |
| Molecular Function: |
UTP:galactose-1-phosphate uridylyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTAEEFDFSS HSHRRYNPLT DSWILVSPHR AKRPWLGQQE AAYKPTAPLY DPKCYLCPGN 60
61 KRATGNLNPR YESTYIFPND YAAVRLDQPI LPQNDSNEDN LKNRLLKVQS VRGNCFVICF 120
121 SPNHNLTIPQ MKQSDLVHIV NSWQALTDDL SREARENHKP FKYVQIFENK GTAMGCSNLH 180
181 PHGQAWCLES IPSEVSQELK SFDKYKREHN TDLFADYVKL ESREKSRVVV ENESFIVVVP 240
241 YWAIWPFETL VISKKKLASI SQFNQMVKED LASILKQLTI KYDNLFETSF PYSMGIHQAP 300
301 LNATGDELSN SWFHMHFYPP LLRSATVRKF LVGFELLGEP QRDLTSEQAA EKLRNLDGQI 360
361 HYLQRL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |

|
ACC1 ADH4 ADR1 AHA1 ARP2 ATP3 AVO1 BCK1 BTN2 BUD7 CBK1 CCT2 CCT3 CCT5 CCT6 CKA1 CLU1 COP1 COR1 CPA2 CPR6 CTR1 DEF1 DHH1 DPB2 EAP1 ECM10 EGD2 FOL2 FUN12 GAL7 GDT1 GND1 GPH1 IDH1 ILV2 ILV5 IPP1 KIC1 KIN2 LHS1 LSC1 LST8 LYS12 MAM33 MDH1 MEH1 MET16 MGE1 MKK2 MKT1 NOG2 NPA3 OYE2 PDC6 PDX1 PHO85 PHO86 PIL1 PMA1 POR1 PRB1 PRE3 PTC7 PUF3 QCR2 RCN2 RPN12 RPN6 RPN7 RPT3 RRP3 SEC27 SEC28 SGT2 SIP1 SIS1 SKG3 SLC1 SLD2 SLT2 SLX1 SMK1 SRP1 SRP54 SSD1 STI1 TAL1 TCP1 TEM1 TFC7 TIF1, TIF2 UBP15 VMA8 YBT1 YCR076C YER077C YGR266W YKR051W YKU80 YLR271W YML020W YPR003C |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
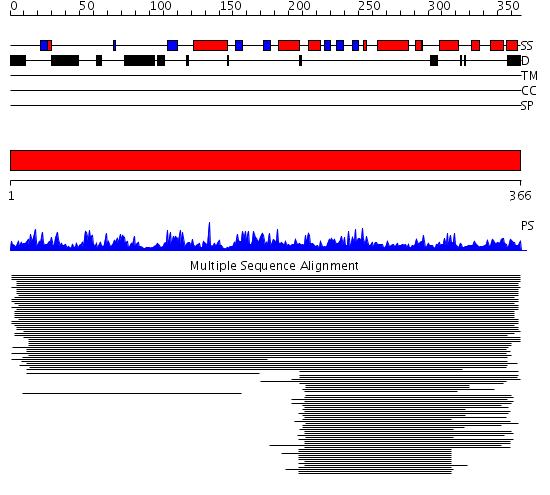
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..366] | 1112.9794 | Galactose-1-phosphate uridylyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)