













| General Information: |
|
| Name(s) found: |
GIP1 /
YBR045C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 573 amino acids |
Gene Ontology: |
|
| Cellular Component: |
protein phosphatase type 1 complex
[IPI prospore membrane [IDA |
| Biological Process: |
regulation of phosphoprotein phosphatase activity
[IMP ascospore wall assembly [IMP |
| Molecular Function: |
protein phosphatase type 1 regulator activity
[IDA protein phosphatase 1 binding [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METILQPKAR PFESLKRKRF REWLRPSTAH GSLLHSDTLD LRDFAKPNPA DTFSNLDSGH 60
61 CPLVTTPIKY ECPDGKSSFF RGDTKFETLF SNRKFYEFKD NLKRGLKKIR HGRNGHQSEK 120
121 RCPVVEETKK SVSDNLDKPD NNTPCFDRFH TNSKEFETQF DHSNRSQNSE KAYLDNESCW 180
181 NLSEKFIPFN NLKYEDLKHF EENLQSLAPA TFTPIESNES LDRSDSTRGT KRSIRNDSSD 240
241 TTSEKRLCLK QYSDEPESDH SMESTPSIYI TKEVQERIEA LSSTDSFLIE KVDFPSNKIG 300
301 SSASDYESDN EYRNMDEDSI NDVTTEKEGN VVIPDSNTST VDAMEKPIEV SSALKDDTLD 360
361 KDIDDASSSY SDDVETTFEP VESEELSDLS DTSSSGSSKI YTIPTFRGLT NRTNISQILS 420
421 KVGKADLSQD NLTHLIKSHQ KKKRCVNFRN KRFYDAFNPY VDNEEDAELS DSENISEMDT 480
481 DLCIKDRSTS SVRFDENSRL LIYKKSKKLN KDETQSGYST TEMRSILKTK MNSQHDEESQ 540
541 RASKCDTVGV AQFLHYFQYT EYKRQRNEAE IID |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
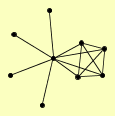
| View Details | Krogan NJ, et al. (2006) |

|
GEP7 GIP1 |
| View Details | Qiu et al. (2008) |
|
ADY3 GAC1 GIP1 GIP3 PIG1 REG1 REG2 SHC1 SPO14 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Uetz P, et al. (2000) |
New Feature: Upload Your Own Microscopy Data
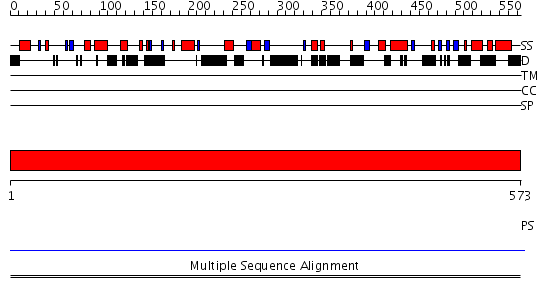
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..573] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)