













| General Information: |
|
| Name(s) found: |
PHO81 /
YGR233C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1178 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
phosphate metabolic process
[TAS |
| Molecular Function: |
cyclin-dependent protein kinase inhibitor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKFGKYLEAR QLELAEYNSH FIDYKALKKL IKQLAIPTLK ASSDLDLHLT LDDIDEKIIH 60
61 QRLQENKAAF FFKLERELEK VNGYYLARES DLRIKFNILH SKYKDYKING KLNSNQATSF 120
121 KNLYAAFKKF QKDLRNLEQY VELNKTGFSK ALKKWDKRSQ SHDKDFYLAT VVSIQPIFTR 180
181 DGPLKLNDET LHILLELNDI DNNNRRADIQ SSTFTNDDDD DNNTSNNNKH NNNNNNNNNN 240
241 NNNNNNNNIL HNNYELTTSK ISENQLEHLF QASSSSLDME MEIENWYKEI LNIATVKDVQ 300
301 RKHALLRNFR ETKIFTYLLQ NSSESFHKNV FSLLKECLTT LFLLLVASPL DDNSLHIFYK 360
361 SNQDHIDLSY CDEDDQVFSR KNVFHEAASC PEKSRLFILD EALTTSKLSK ETVQKLLNAQ 420
421 DIHSRVPLHY AAELGKLEFV HSLLITNLLE DVDPIDSDSK TPLVLAITNN HIDVVRDLLT 480
481 IGGANASPIE KPILDYSKNV ISSTKVQFDP LNVACKFNNH DAAKLLLEIR SKQNADNAKN 540
541 KSSQHLCQPL FKKNSTGLCT LHIVAKIGGD PQLIQLLIRY GADPNEIDGF NKWTPIFYAV 600
601 RSGHSEVITE LLKHNARLDI EDDNGHSPLF YALWESHVDV LNALLQRPLN LPSAPLNEIN 660
661 SQSSTQRLNT IDLTPNDDKF DLDIQDSIPD FALPPPIIPL RKYGHNFLEK KIFIKLKLRP 720
721 GLESIKLTQD NGIIMSSSPG RITLSSNLPE IIPRNVILPV RSGEINNFCK DISETNDEED 780
781 DDEISEDHDD GEIIFQVDSI DDFSMDFEIF PSFGTRIIAK TTAMPFLFKK VAINSIATMN 840
841 LPLFDTRLNN IGSLTLDYQI IFPYPGNPLK IINYEPYWKS TGSDLMTSSK DGNFVTSSSL 900
901 NGSFISVLVC ALNDETIVAA PKPYVEFKGT KILLNDLTKE QLEKVVDYDF GKIDGSFDEV 960
961 TLKQYLSSRV VPLRSLLEVI PGSAQLVIRV YFPTDKEIDT IPIKISPFIN INQFIDKLLL1020
1021 IIFEHERFLR HSGSGSMRQI VFSSCNWEAC SILNWKQPNF PVLLQMKNLL RDSTTGKFVG1080
1081 DTPNCLKELA VNPQKMSYLN TELINIHTMV QFAMNNNLLG VTLPYEVLKI CPSLARIIKQ1140
1141 NGLLLIASVG ENDQIPADGG YSGIYYACEL LFENNIDM |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
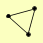
|
PHO80 PHO81 PHO85 |
| View Details | Krogan NJ, et al. (2006) |

|
PHO80 PHO81 |
| View Details | Gavin AC, et al. (2006) |
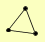
|
CYR1 PHO81 SRV2 |
| View Details | Gavin AC, et al. (2002) |
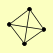
|
ACT1 CYR1 PHO81 SRV2 |
| View Details | Ho Y, et al. (2002) |
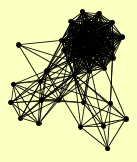
|
AAC3 ADK1 AGX1 AIP1 BUD32 CAR2 COF1 CPR6 CRM1 CYR1 DIA4 EMG1 GCN1 GRX3 GRX4 HEF3 IDP2 IMD2 IMD4 KAE1 KAP114 NPA3 PCL7 PHO81 PHO85 REX2 RNH202 RPN1 RPN5 RPN6 RPT1 SEC18 SEC23 SGO1 SOD1 SRV2 TMA29 TOS3 URA7 YHB1 YHR033W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | experimental1 - mih1-1 | McCusker D, et al (2007) | |
| View Run | 3906: TAP-tagged | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
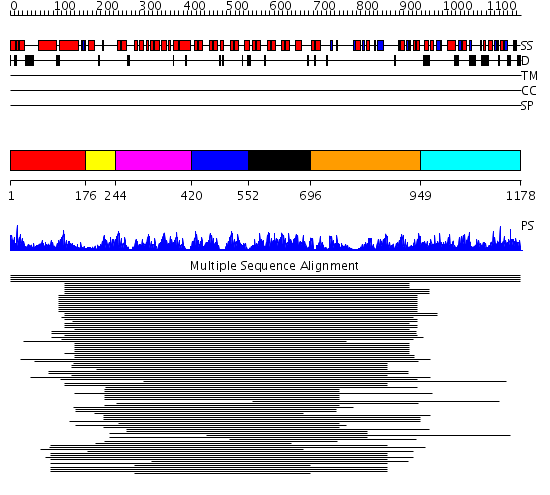
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..175] | 53.585027 | SPX domain No confident structure predictions are available. | |
| 2 | View Details | [176..243] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [244..419] | 7.62 | Hemolysin E (HlyE, ClyA, SheA) | |
| 4 | View Details | [420..551] | 109.228787 | bcl-3 | |
| 5 | View Details | [552..695] | 109.228787 | bcl-3 | |
| 6 | View Details | [696..948] | 1.463992 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [949..1178] | 8.053994 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)