













| General Information: |
|
| Name(s) found: |
STH1 /
YIL126W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1359 amino acids |
Gene Ontology: |
|
| Cellular Component: |
RSC complex
[IDA |
| Biological Process: |
cytoskeleton organization
[IMP ATP-dependent chromatin remodeling [IDA chromosome segregation [IGI chromatin remodeling at centromere [IMP meiosis [IMP double-strand break repair [IMP |
| Molecular Function: |
ATPase activity
[IDA DNA-dependent ATPase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLQEQSELMS TVMNNTPTTV AALAAVAAAS ETNGKLGSEE QPEITIPKPR SSAQLEQLLY 60
61 RYRAIQNHPK ENKLEIKAIE DTFRNISRDQ DIYETKLDTL RKSIDKGFQY DEDLLNKHLV 120
121 ALQLLEKDTD VPDYFLDLPD TKNDNTTAIE VDYSEKKPIK ISADFNAKAK SLGLESKFSN 180
181 ATKTALGDPD TEIRISARIS NRINELERLP ANLGTYSLDD CLEFITKDDL SSRMDTFKIK 240
241 ALVELKSLKL LTKQKSIRQK LINNVASQAH HNIPYLRDSP FTAAAQRSVQ IRSKVIVPQT 300
301 VRLAEELERQ QLLEKRKKER NLHLQKINSI IDFIKERQSE QWSRQERCFQ FGRLGASLHN 360
361 QMEKDEQKRI ERTAKQRLAA LKSNDEEAYL KLLDQTKDTR ITQLLRQTNS FLDSLSEAVR 420
421 AQQNEAKILH GEEVQPITDE EREKTDYYEV AHRIKEKIDK QPSILVGGTL KEYQLRGLEW 480
481 MVSLYNNHLN GILADEMGLG KTIQSISLIT YLYEVKKDIG PFLVIVPLST ITNWTLEFEK 540
541 WAPSLNTIIY KGTPNQRHSL QHQIRVGNFD VLLTTYEYII KDKSLLSKHD WAHMIIDEGH 600
601 RMKNAQSKLS FTISHYYRTR NRLILTGTPL QNNLPELWAL LNFVLPKIFN SAKTFEDWFN 660
661 TPFANTGTQE KLELTEEETL LIIRRLHKVL RPFLLRRLKK EVEKDLPDKV EKVIKCKLSG 720
721 LQQQLYQQML KHNALFVGAG TEGATKGGIK GLNNKIMQLR KICNHPFVFD EVEGVVNPSR 780
781 GNSDLLFRVA GKFELLDRVL PKFKASGHRV LMFFQMTQVM DIMEDFLRMK DLKYMRLDGS 840
841 TKTEERTEML NAFNAPDSDY FCFLLSTRAG GLGLNLQTAD TVIIFDTDWN PHQDLQAQDR 900
901 AHRIGQKNEV RILRLITTDS VEEVILERAM QKLDIDGKVI QAGKFDNKST AEEQEAFLRR 960
961 LIESETNRDD DDKAELDDDE LNDTLARSAD EKILFDKIDK ERMNQERADA KAQGLRVPPP1020
1021 RLIQLDELPK VFREDIEEHF KKEDSEPLGR IRQKKRVYYD DGLTEEQFLE AVEDDNMSLE1080
1081 DAIKKRREAR ERRRLRQNGT KENEIETLEN TPEASETSLI ENNSFTAAVD EETNADKETT1140
1141 ASRSKRRSSR KKRTISIVTA EDKENTQEES TSQENGGAKV EEEVKSSSVE IINGSESKKK1200
1201 KPKLTVKIKL NKTTVLENND GKRAEEKPES KSPAKKTAAK KTKTKSKSLG IFPTVEKLVE1260
1261 EMREQLDEVD SHPRTSIFEK LPSKRDYPDY FKVIEKPMAI DIILKNCKNG TYKTLEEVRQ1320
1321 ALQTMFENAR FYNEEGSWVY VDADKLNEFT DEWFKEHSS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ARP7 ARP9 HTB1 HTL1 ISW1 ISW2 ITC1 MOT1 NFI1 NPL6 RSC1 RSC2 RSC3 RSC30 RSC4 RSC58 RSC6 RSC8 RSC9 RTT102 SFH1 STH1 VPS1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
ARP7 ARP9 RSC1 RSC2 RSC4 RSC6 RSC8 SAS5 SFH1 STH1 |
| View Details | Krogan NJ, et al. (2006) |

|
ARP7 ARP9 ATG16 CSH1 GRS2 HTL1 MGR1 NFI1 NPL6 PCL9 RIM15 RSC1 RSC2 RSC3 RSC4 RSC58 RSC6 RSC8 RSC9 RTT102 SFH1 STH1 URC2 YIL177C |
| View Details | Gavin AC, et al. (2006) |
|
ARP7 NPL6 RSC1 RSC2 RSC3 RSC4 RSC58 RSC6 RSC8 RSC9 SFH1 STH1 |
| View Details | Gavin AC, et al. (2002) |

|
ADE5,7 DPB2 DPB4 HHF1, HHF2 IOC3 ISW1 ISW2 ITC1 KAP114 MOT1 NAP1 NPL6 POL2 PUF6 REB1 RET1 RFA1 RFX1 RNR2 RPA135 RPC34 RPC40 RPC82 RPO31 RSC2 RSC3 RSC58 RSC8 RVB2 SFH1 STH1 STO1 TAF1 TFC7 TOP1 VPS1 YBR090C-A, NHP6B |
| View Details | Ho Y, et al. (2002) |
|
DMA2 GDE1 PHO84 STH1 |
| View Details | Qiu et al. (2008) |
|
ARP7 ARP9 HTL1 RSC4 RSC58 RSC6 RSC8 SAS5 SFH1 SNF5 STH1 TAF14 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | control 1 | McCusker D, et al (2007) | |
| View Run | control 2 | McCusker D, et al (2007) | |
| View Run | experimental1 - mih1-1 | McCusker D, et al (2007) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
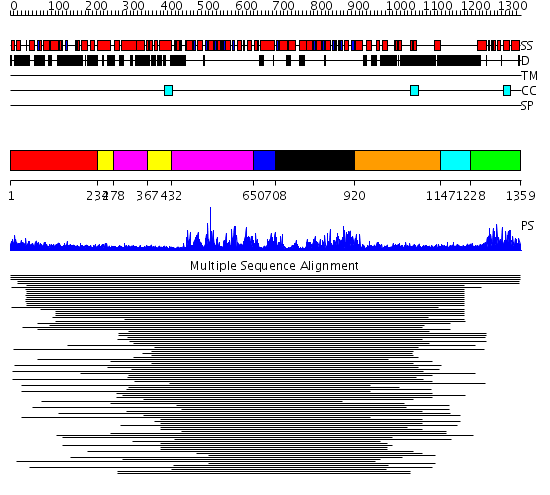
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..233] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [234..277] [367..431] |
4.0 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 3 | View Details | [278..366] [432..649] |
4.0 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 4 | View Details | [650..707] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [708..919] | 3.0 | Putative DEAD box RNA helicase | |
| 6 | View Details | [920..1146] | 3.390994 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1147..1227] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1228..1359] | 72.522879 | GCN5 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.46 |
Source: Reynolds et al. (2008)