













| General Information: |
|
| Name(s) found: |
RSC1 /
YGR056W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 928 amino acids |
Gene Ontology: |
|
| Cellular Component: |
RSC complex
[IDA |
| Biological Process: |
ATP-dependent chromatin remodeling
[IC double-strand break repair via nonhomologous end joining [IPI |
| Molecular Function: |
DNA-dependent ATPase activity
[IC |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVEQDNGFLQ KLLKTQYDAV FHLKDENGIE IYPIFNVLPP KKEYPDYYII IRNPISLNTL 60
61 KKRLPHYTSP QDFVNDFAQI PWNAMTYNAK DSVIYKYAIL LESFIKGKIV HNIRKHYPEV 120
121 TYPSLGRIPE IFAESMQPSD LSSNPINTQE NDEKAGLNPE MKMAFAKLDS SITERKPTNQ 180
181 DYRMQQKNSP AFPTHSASIT PQPLASPTPV VNYANITSAH PKTHVRRGRP PVIDLPYVLR 240
241 IKNILKMMRR EVDQNNKTLT LCFEKLPDRN EEPTYYSVIT DPICLMDIRK KVKSRKYRNF 300
301 HTFEEDFQLM LTNFKLYYSQ DQSNIIRAQL LEKNFNRLVR IELSKPDEDY LPEGELRYPL 360
361 DDVEINDEKY QIGDWVLLHN PNDINKPIVG QIFRLWSTTD GNKWLNACWY FRPEQTVHRV 420
421 DRLFYKNEVM KTGQYRDHPI QDIKGKCYVI HFTRFQRGDP STKVNGPQFV CEFRYNESDK 480
481 VFNKIRTWKA CLPEELRDQD EPTIPVNGRK FFKYPSPIAD LLPANATLND KVPEPTEGAP 540
541 TAPPLVGAVY LGPKLERDDL GEYSTSDDCP RYIIRPNDPP EEGKIDYETG TIITDTLTTS 600
601 SMPRVNSSST IRLPTLKQTK SIPSSNFRSS SNTPLLHQNF NQTSNFLKLE NMNNSSHNLL 660
661 SHPSVPKFQS PSLLEQSSRS KYHSAKKQTQ LSSTAPKKPA SKSFTLSSMI NTLTAHTSKY 720
721 NFNHIVIEAP GAFVVPVPME KNIRTIQSTE RFSRSNLKNA QNLGNTAIND INTANEQIIW 780
781 FKGPGVKITE RVIDSGNDLV RVPLNRWFCK NKRRKLDYED IEEDVMEPPN DFSEDMIANI 840
841 FNPPPSLNLD MDLNLSPSSN NSSNFMDLST IASGDNDGKE CDTAEESEDE NEDTEDEHEI 900
901 EDIPTTSAFG LNSSAEYLAF RLREFNKL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ARP7 ARP9 NPL6 RSC1 RSC2 RSC3 RSC30 RSC4 RSC58 RSC6 RSC8 RSC9 RTT102 SFH1 STH1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
ARP7 ARP9 RSC1 RSC2 RSC4 RSC6 RSC8 SAS5 SFH1 STH1 |
| View Details | Krogan NJ, et al. (2006) |

|
ARP7 ARP9 ATG16 CSH1 GRS2 HTL1 MGR1 NFI1 NPL6 PCL9 RIM15 RSC1 RSC2 RSC3 RSC4 RSC58 RSC6 RSC8 RSC9 RTT102 SFH1 STH1 URC2 YIL177C |
| View Details | Gavin AC, et al. (2006) |
|
ARP7 NPL6 RSC1 RSC2 RSC3 RSC4 RSC58 RSC6 RSC8 RSC9 SFH1 STH1 |
| View Details | Qiu et al. (2008) |
|
ARP7 ARP9 RSC1 RSC2 RSC30 RSC4 RSC6 RSC8 SFH1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
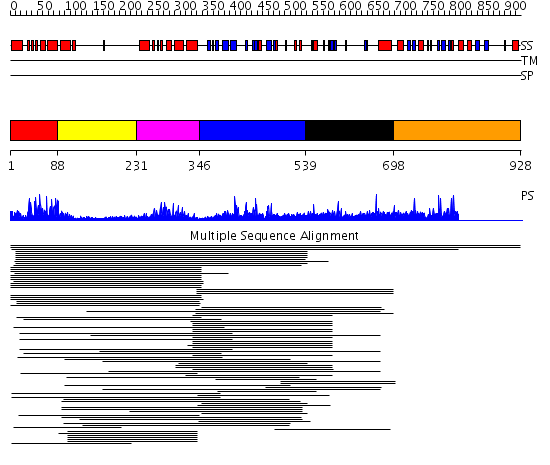
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | 69.751666 | GCN5 | |
| 2 | View Details | [88..230] | 46.917458 | TAFII250 double bromodomain module | |
| 3 | View Details | [231..345] | 46.917458 | TAFII250 double bromodomain module | |
| 4 | View Details | [346..538] | 7.88 | Origin-recognition complect protein 120kDa subunit, Orc1p | |
| 5 | View Details | [539..697] | 1.034996 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [698..928] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)