













| General Information: |
|
| Name(s) found: |
OSH3 /
YHR073W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 996 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
sterol transport
[IGI pseudohyphal growth [IMP invasive growth in response to glucose limitation [IGI karyogamy involved in conjugation with cellular fusion [IGI endocytosis [IGI exocytosis [IGI |
| Molecular Function: |
oxysterol binding
[ISS phosphatidylinositol binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METIDIQNRS FVVRWVKCGR GDVINYQIKP LKKSIEVGIY KKLKSSVDDH ASAVHIAPDT 60
61 KTLLDYTTKS LLHKGSSSNI EEHHRRSSQH SHSSSNGSDN KRKERSYSSL SISGIQQQSQ 120
121 EIPLREKLSA SGFTLVKRVG NVSGNTMVQG DLEVKDTDYY YAFILDNSSS KNAKKKILFN 180
181 ASVINGDNQS MISTRSTPPA RPTALSRTST QQDMLFRVGQ GRYLQGYLLK KRRKRLQGFK 240
241 KRFFTLDFRY GTLSYYLNDH NQTCRGEIVI SLSSVSANKK DKIIIIDSGM EVWVLKATTK 300
301 ENWQSWVDAL QTCFDDQFED KDTSTLEENP DILDDDKEVI NKSSPQDHDH LTPTATTKSA 360
361 LSHRQHTQKD MDDIYVPLPS ESYATFSMNL RLIQQRLEQC KKDSLSYKPT TLHQRSEGLN 420
421 GTHSSSSVFT NNRVSSFNHS SSGMTSSDSL ASEEVPSNKT YIEHALYNQL ADLEVFVSRF 480
481 VTQGEVLFKD HQILCKKAKD TRVSLTSYLS ENDEFFDAEE EISRGVIILP DTEDDINNIV 540
541 EETPLLGKSD QNEFTKEVQL SGSEQIASSS VESYTTNDEN HSRKHLKNRH KNRRRGHPHH 600
601 QKTKSAQSST ETFTSKDLFA LSYPKSVTRR NDIPEAAASP PSLLSFLRKN VGKDLSSIAM 660
661 PVTSNEPISI LQLISETFEY APLLTKATQR PDPITFVSAF AISFLSIYRD KTRTLRKPFN 720
721 PLLAETFELI REDMGFRLIS EKVSHRPPVF AFFAEHLDWE CSYTVTPSQK FWGKSIELNN 780
781 EGILRLKFKT TGELFEWTQP TTILKNLIAG ERYMEPVNEF EVHSSKGDKS HILFDKAGMF 840
841 SGRSEGFKVS IIPPPSSNRK KETLAGKWTQ SLANETTHET IWEVGDLVSN PKKKYGFTKF 900
901 TANLNEITEI EKGNLPPTDS RLRPDIRAYE EGNVDKAEEW KLKLEQLQRE RRNKGQDVEP 960
961 KYFEKVSKNE WKYITGPKSY WERRKKHDWS DISQLW |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
COR1 OSH3 QCR2 |
| View Details | Ho Y, et al. (2002) |
|
NOP4 OSH3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
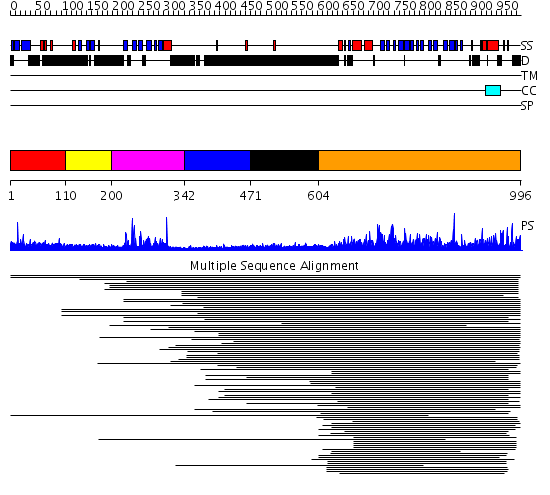
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | 23.69897 | Pleckstrin, N-terminal domain | |
| 2 | View Details | [110..199] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [200..341] | 16.522879 | Insulin receptor substrate 1, IRS-1 | |
| 4 | View Details | [342..470] | 16.522879 | Insulin receptor substrate 1, IRS-1 | |
| 5 | View Details | [471..603] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [604..996] | 13.089929 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)