













| General Information: |
|
| Name(s) found: |
UBR1 /
YGR184C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1950 amino acids |
Gene Ontology: |
|
| Cellular Component: |
proteasome complex
[IPI |
| Biological Process: |
protein polyubiquitination
[TAS protein monoubiquitination [TAS |
| Molecular Function: |
ubiquitin-protein ligase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVADDDLGS LQGHIRRTLR SIHNLPYFRY TRGPTERADM SRALKEFIYR YLYFVISNSG 60
61 ENLPTLFNAH PKQKLSNPEL TVFPDSLEDA VDIDKITSQQ TIPFYKIDES RIGDVHKHTG 120
121 RNCGRKFKIG EPLYRCHECG CDDTCVLCIH CFNPKDHVNH HVCTDICTEF TSGICDCGDE 180
181 EAWNSPLHCK AEEQENDISE DPATNADIKE EDVWNDSVNI ALVELVLAEV FDYFIDVFNQ 240
241 NIEPLPTIQK DITIKLREMT QQGKMYERAQ FLNDLKYEND YMFDGTTTAK TSPSNSPEAS 300
301 PSLAKIDPEN YTVIIYNDEY HNYSQATTAL RQGVPDNVHI DLLTSRIDGE GRAMLKCSQD 360
361 LSSVLGGFFA VQTNGLSATL TSWSEYLHQE TCKYIILWIT HCLNIPNSSF QTTFRNMMGK 420
421 TLCSEYLNAT ECRDMTPVVE KYFSNKFDKN DPYRYIDLSI LADGNQIPLG HHKILPESST 480
481 HSLSPLINDV ETPTSRTYSN TRLQHILYFD NRYWKRLRKD IQNVIIPTLA SSNLYKPIFC 540
541 QQVVEIFNHI TRSVAYMDRE PQLTAIRECV VQLFTCPTNA KNIFENQSFL DIVWSIIDIF 600
601 KEFCKVEGGV LIWQRVQKSN LTKSYSISFK QGLYTVETLL SKVHDPNIPL RPKEIISLLT 660
661 LCKLFNGAWK IKRKEGEHVL HEDQNFISYL EYTTSIYSII QTAEKVSEKS KDSIDSKLFL 720
721 NAIRIISSFL GNRSLTYKLI YDSHEVIKFS VSHERVAFMN PLQTMLSFLI EKVSLKDAYE 780
781 ALEDCSDFLK ISDFSLRSVV LCSQIDVGFW VRNGMSVLHQ ASYYKNNPEL GSYSRDIHLN 840
841 QLAILWERDD IPRIIYNILD RWELLDWFTG EVDYQHTVYE DKISFIIQQF IAFIYQILTE 900
901 RQYFKTFSSL KDRRMDQIKN SIIYNLYMKP LSYSKLLRSV PDYLTEDTTE FDEALEEVSV 960
961 FVEPKGLADN GVFKLKASLY AKVDPLKLLN LENEFESSAT IIKSHLAKDK DEIAKVVLIP1020
1021 QVSIKQLDKD ALNLGAFTRN TVFAKVVYKL LQVCLDMEDS TFLNELLHLV HGIFRDDELI1080
1081 NGKDSIPEAY LSKPICNLLL SIANAKSDVF SESIVRKADY LLEKMIMKKP NELFESLIAS1140
1141 FGNQYVNDYK DKKLRQGVNL QETEKERKRR LAKKHQARLL AKFNNQQTKF MKEHESEFDE1200
1201 QDNDVDMVGE KVYESEDFTC ALCQDSSSTD FFVIPAYHDH SPIFRPGNIF NPNEFMPMWD1260
1261 GFYNDDEKQA YIDDDVLEAL KENGSCGSRK VFVSCNHHIH HNCFKRYVQK KRFSSNAFIC1320
1321 PLCQTFSNCT LPLCQTSKAN TGLSLDMFLE SELSLDTLSR LFKPFTEENY RTINSIFSLM1380
1381 ISQCQGFDKA VRKRANFSHK DVSLILSVHW ANTISMLEIA SRLEKPYSIS FFRSREQKYK1440
1441 TLKNILVCIM LFTFVIGKPS MEFEPYPQQP DTVWNQNQLF QYIVRSALFS PVSLRQTVTE1500
1501 ALTTFSRQFL RDFLQGLSDA EQVTKLYAKA SKIGDVLKVS EQMLFALRTI SDVRMEGLDS1560
1561 ESIIYDLAYT FLLKSLLPTI RRCLVFIKVL HELVKDSENE TLVINGHEVE EELEFEDTAE1620
1621 FVNKALKMIT EKESLVDLLT TQESIVSHPY LENIPYEYCG IIKLIDLSKY LNTYVTQSKE1680
1681 IKLREERSQH MKNADNRLDF KICLTCGVKV HLRADRHEMT KHLNKNCFKP FGAFLMPNSS1740
1741 EVCLHLTQPP SNIFISAPYL NSHGEVGRNA MRRGDLTTLN LKRYEHLNRL WINNEIPGYI1800
1801 SRVMGDEFRV TILSNGFLFA FNREPRPRRI PPTDEDDEDM EEGEDGFFTE GNDEMDVDDE1860
1861 TGQAANLFGV GAEGIAGGGV RDFFQFFENF RNTLQPQGNG DDDAPQNPPP ILQFLGPQFD1920
1921 GATIIRNTNP RNLDEDDSDD NDDSDEREIW |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |

|
AHA1 AHP1 ARF1 CCR4 CCT2 CCT3 CCT5 CCT6 CDC36 CDC39 COP1 CYS4 DCS1 DCS2 FZO1 GDB1 GFA1 GND1 GPG1 GSY1 HEF3 HTB1 LHS1 MGE1 MGT1 MYO2 NEW1 NUG1 PEX7 PIL1 POP2 RAD50 RFC4 RIP1 RNQ1 RPN6 RVB1 SEC27 SMC3 SOF1 STI1 TMA19 TSA2 UBP15 UBR1 URA7 VPS1 YAK1 YGL081W YHR033W YLR241W YPL247C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample 3121 run on DecaLCQ: searched for phosphorylation | Green EM, et al (2005) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
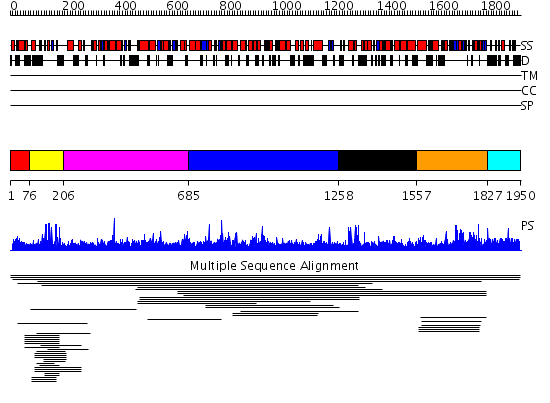
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [76..205] | 35.853872 | Putative zinc finger in N-recognin No confident structure predictions are available. | |
| 3 | View Details | [206..684] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [685..1257] | 2.0159 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1258..1556] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1557..1826] | 5.00995 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1827..1950] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [871..1009] | 2.35 | No description for 2doaA was found. | |
| 9 | View Details | [1010..1156] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1157..1327] | 2.060995 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1328..1554] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1555..1654] | 1.044991 | View MSA. No confident structure predictions are available. | |
| 13 | View Details | [1655..1805] | 3.049987 | View MSA. No confident structure predictions are available. | |
| 14 | View Details | [1806..1950] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 8 |
|
|||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 13 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 14 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)