













| General Information: |
|
| Name(s) found: |
CCR4 /
YAL021C
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 837 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA Cdc73/Paf1 complex [IPI CCR4-NOT core complex [IPI |
| Biological Process: |
response to drug
[IMP traversing start control point of mitotic cell cycle [IMP nuclear-transcribed mRNA poly(A) tail shortening [IDA nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay [IDA regulation of transcription from RNA polymerase II promoter [IPI RNA elongation from RNA polymerase II promoter [IGI |
| Molecular Function: |
3'-5'-exoribonuclease activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNDPSLLGYP NVGPQQQQQQ QQQQHAGLLG KGTPNALQQQ LHMNQLTGIP PPGLMNNSDV 60
61 HTSSNNNSRQ LLDQLANGNA NMLNMNMDNN NNNNNNNNNN NNNGGGSGVM MNASTAAVNS 120
121 IGMVPTVGTP VNINVNASNP LLHPHLDDPS LLNNPIWKLQ LHLAAVSAQS LGQPNIYARQ 180
181 NAMKKYLATQ QAQQAQQQAQ QQAQQQVPGP FGPGPQAAPP ALQPTDFQQS HIAEASKSLV 240
241 DCTKQALMEM ADTLTDSKTA KKQQPTGDST PSGTATNSAV STPLTPKIEL FANGKDEANQ 300
301 ALLQHKKLSQ YSIDEDDDIE NRMVMPKDSK YDDQLWHALD LSNLQIFNIS ANIFKYDFLT 360
361 RLYLNGNSLT ELPAEIKNLS NLRVLDLSHN RLTSLPAELG SCFQLKYFYF FDNMVTTLPW 420
421 EFGNLCNLQF LGVEGNPLEK QFLKILTEKS VTGLIFYLRD NRPEIPLPHE RRFIEINTDG 480
481 EPQREYDSLQ QSTEHLATDL AKRTFTVLSY NTLCQHYATP KMYRYTPSWA LSWDYRRNKL 540
541 KEQILSYDSD LLCLQEVESK TFEEYWVPLL DKHGYTGIFH AKARAKTMHS KDSKKVDGCC 600
601 IFFKRDQFKL ITKDAMDFSG AWMKHKKFQR TEDYLNRAMN KDNVALFLKL QHIPSGDTIW 660
661 AVTTHLHWDP KFNDVKTFQV GVLLDHLETL LKEETSHNFR QDIKKFPVLI CGDFNSYINS 720
721 AVYELINTGR VQIHQEGNGR DFGYMSEKNF SHNLALKSSY NCIGELPFTN FTPSFTDVID 780
781 YIWFSTHALR VRGLLGEVDP EYVSKFIGFP NDKFPSDHIP LLARFEFMKT NTGSKKV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
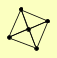
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CAF130 CAF40 CCR4 CDC39 POP2 |
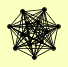
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
CAF16 CAF4 CCR4 CDC36 CDC39 DBF2 DHH1 IBA57 MOB1 MOT2 NOT3 NOT5 POP2 |
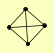
| View Details | Krogan NJ, et al. (2006) |
|
CAF40 CCR4 CDC39 POP2 |
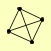
| View Details | Gavin AC, et al. (2006) |
|
CAF40 CCR4 CDC39 POP2 |
| View Details | Gavin AC, et al. (2002) |

|
AHA1 ATP11 CAF130 CAF40 CCR4 CCT6 CDC36 CDC39 COP1 FAS2 GCN1 MOT2 NOT3 NOT5 PDC1 POL1 POL12 POP2 PRI1 PRI2 RVB2 SAM1 SEC27 TFC7 TFP1 YRA1 |
| View Details | Ho Y, et al. (2002) |
|
CCR4 CDC36 CDC39 CYS4 PIL1 POP2 RNQ1 RVB1 STI1 UBR1 |
| View Details | Qiu et al. (2008) |
|
CAF16 CAF4 CAF40 CCR4 CDC36 CDC39 DBF2 DHH1 IBA57 MOB1 MOT2 NOT3 NOT5 POP2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
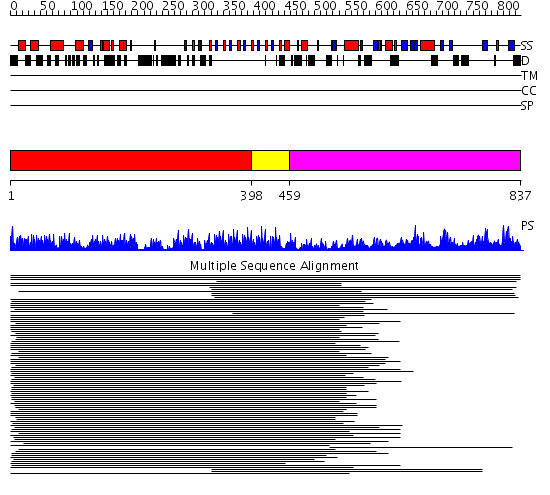
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..397] | 16.221849 | Leucine rich effector protein YopM | |
| 2 | View Details | [398..458] | 13.69897 | No description for 1k14A_ was found. | |
| 3 | View Details | [459..837] | 14.32 | DNA repair endonuclease Hap1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)