













| General Information: |
|
| Name(s) found: |
CAF16 /
YFL028C
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 289 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA CCR4-NOT complex [IPI |
| Biological Process: |
regulation of transcription, DNA-dependent
[IMP |
| Molecular Function: |
ATPase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVSQFAIEVR NLTYKFKESS DPSVVDINLQ IPWNTRSLVV GANGAGKSTL LKLLSGKHLC 60
61 LDGKILVNGL DPFSPLSMNQ VDDDESVEDS TNYQTTTYLG TEWCHMSIIN RDIGVLELLK 120
121 SIGFDHFRER GERLVRILDI DVRWRMHRLS DGQKRRVQLA MGLLKPWRVL LLDEVTVDLD 180
181 VIARARLLEF LKWETETRRC SVVYATHIFD GLAKWPNQVY HMKSGKIVDN LDYQKDVEFS 240
241 EVVNAKVNGQ VAFENDNNKV VISKVNSLHP LALEWLKRDN QIPDKEIGI |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
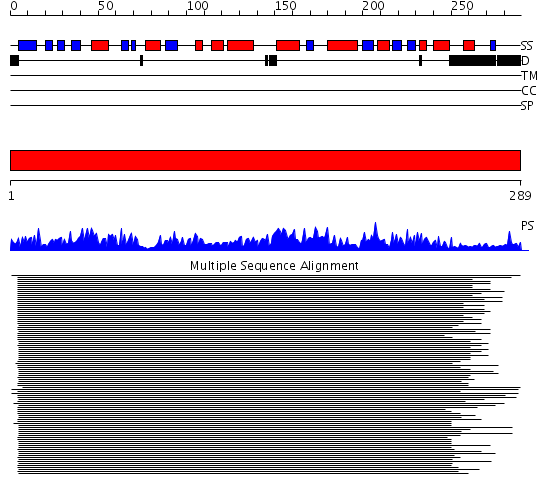
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..289] | 179.071898 | MsbA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)