













| General Information: |
|
| Name(s) found: |
SRY1 /
YKL218C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 326 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
cellular amino acid derivative catabolic process
[IMP |
| Molecular Function: |
threo-3-hydroxyaspartate ammonia-lyase activity
[IDA [NOT]serine racemase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIVPTYGDVL DASNRIKEYV NKTPVLTSRM LNDRLGAQIY FKGENFQRVG AFKFRGAMNA 60
61 VSKLSDEKRS KGVIAFSSGN HAQAIALSAK LLNVPATIVM PEDAPALKVA ATAGYGAHII 120
121 RYNRYTEDRE QIGRQLAAEH GFALIPPYDH PDVIAGQGTS AKELLEEVGQ LDALFVPLGG 180
181 GGLLSGSALA ARSLSPGCKI FGVEPEAGND GQQSFRSGSI VHINTPKTIA DGAQTQHLGE 240
241 YTFAIIRENV DDILTVSDQE LVKCMHFLAE RMKVVVEPTA CLGFAGALLK KEELVGKKVG 300
301 IILSGGNVDM KRYATLISGK EDGPTI |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
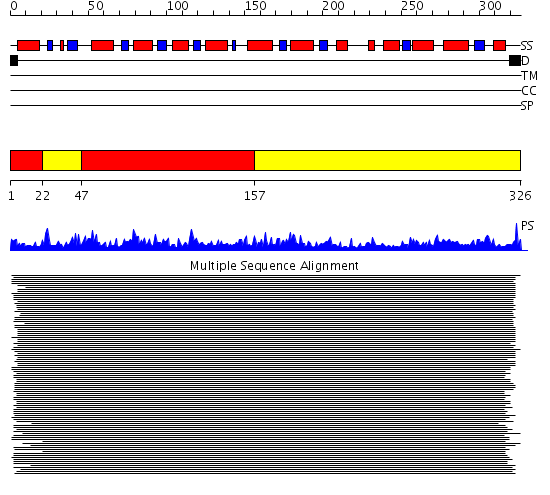
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..21] [47..156] |
506.67837 | Allosteric threonine deaminase N-terminal domain; Allosteric threonine deaminase C-terminal domain | |
| 2 | View Details | [22..46] [157..326] |
506.67837 | Allosteric threonine deaminase N-terminal domain; Allosteric threonine deaminase C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)