













| General Information: |
|
| Name(s) found: |
DOP1 /
YDR141C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1698 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA |
| Biological Process: |
cell morphogenesis
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLPLKPLTI DSNNKQLDSK QKKFRANVER ALERFDSVTE WADYIASLGT LLKALQSWSP 60
61 KFQNVRYYVP SPYQVSRRLT SSLSPALPAG VHQKTLEVYT YIFEHIGLET LATECNIWIP 120
121 GILPLMTYAS MSVRSHLIEL YDNYILLLPQ TTLRLLIRPL ISSLLPGIDD ESNDFLPLTL 180
181 KLIETLQENL DDDSLFWQTL FLVMTANKGR RLGGLTWLTR KFPSLNAVPH LVNKIKMEAE 240
241 ENPSETETND SHLDRKKRKE EAFKVLLPAA KDLVTPEPGL LIRCLVGCLE DENDILIKRS 300
301 VLDLLLQRLR LDSPVLNVLI TSEDKKLLIM SCCRTTLSKD MSLNRRIWNW LLGPTAGGML 360
361 NNNGGNSMEY TTSVKSANEE SNVYFTKYGL SALLEGLSDL LSEEESVLTA FRISMAVMDR 420
421 WEIGSLVIPE LFIPLLYSSE KFKQNEQIMK TARTFFDNTE TNIIWGKLFQ ELEDIKNLKI 480
481 LDFVLTNFNI GNDEEIIVRH LPLILLTLLA LPSNDKDFDN IYKLQKFSLY NKLLNYIPER 540
541 ALLPLSHSKL KHDDEVSCEE LLAKIRGFYT NVSNPSSILE KENIAERLPP FTTEDLTFLI 600
601 ADLIQKKLLS SLWDLENINE SSKLFIAIFE KIPESEELKG RSHISWSDKK ITQSIFEAIP 660
661 RLCESNNDAK SEEIVGIVEI FGNYLYSRME FIESMKLLKV VMMAVWKSLK DPRHQILGVK 720
721 NLKTLNRFIP SKFIESALVY TFVEEEDISE RLSVLDLLWT QLDSDSNLIR RPLELILGEL 780
781 FDDQNPFYLT VSKWILSILN SGSASRLFYI LTDNILKVNR LEKERLDERD DLDMLTYEFQ 840
841 MLAYVLKTNN GRTRKVFSTE LTSIKSSTIW KNEDVSTYKS LLLVTLMRFL NIKSNTHAKS 900
901 IRSALILLDI LLDGTEQNFK DIVIFLLQMS SKYIAEEGIE PELIAVSLLD IVSKVLRLSH 960
961 DNGIKLDIFD DNAAHLKYID FLVTSVSNMK SPLIVTAYVK LLSESIVYFE NSIFRMILPL1020
1021 SASLVQCVQR LFLLEKREGG YYQPIALLLG GLEELLEISH GYLVTEEREG YFSGSNLKGD1080
1081 FIQSVVSNVF SSDSSNEESK IQGERDVILQ SFRQVISCCL DIWYWAHNIS CKSNDDSSLD1140
1141 ATNHNSYKFK FRSKKLLETL FLLEPLELLE NLISIRSDNT TVTLVHVLDG NKPAITIPHL1200
1201 LYGVIIRYNR TASVKFSNRD GSRSSTTKLT KGEPSMLKRL SGESIIAFLF NYVDSVENSA1260
1261 MEEFYGDFLL FFREVATNYN LYSDVSLSIL KLVALISGKV SKTQFGEQKR VRREISDVFF1320
1321 KYLPNAFINF TNLYRGHPDS FKDLEFVVWR VQYIVNDQIG GDKFNTTLAT IVNQCLTPYI1380
1381 KPKSEKTIPG YVLELAAVVS HLGSKVKSWR LLIAELFQND KKLSVIGSDQ TWEKIIYEWS1440
1441 IYPENKSKIL NDLLLEIGSK RSSVTPTLIT FNLGSDSEVE YKCQNLLKIS YLLMVSPNDA1500
1501 YLLHFSSLIS CIFHYLVSKD IKLKGSCWIL LRVLLLRFSE SHFNDYWSMI SYCLQTNLQE1560
1561 FYESLQIQSE VDPQTILQVC KTLDLLLLLN MEGFTSTNEW IFVIDTINCV YKTNSFVALV1620
1621 DEIAEFKDYE ITKTDDLELP TTLKDGLPLL RGIHKIERHT QLRSFFQNLS YLHYEKVYGL1680
1681 GSVDLYGCGE DLKKDILS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
DOP1 MON2 |
| View Details | Gavin AC, et al. (2006) |

|
DOP1 MON2 |
| View Details | Ho Y, et al. (2002) |
|
ADR1 BEM3 DOP1 ECM29 KAP122 MET18 RPA135 RPA190 RPC40 RPO31 SRY1 YER138C YIL055C YTA7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
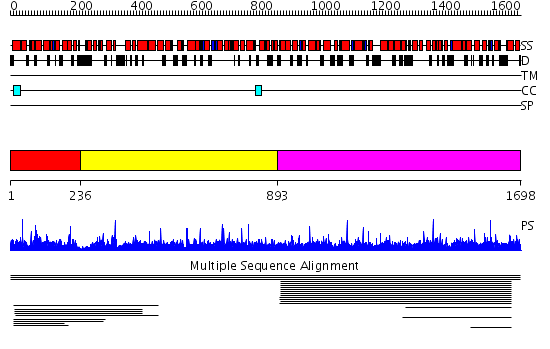
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..235] | 2.009951 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [236..892] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [893..1698] | 4.009001 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [547..668] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [669..892] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [893..962] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [963..1175] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1176..1324] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1325..1541] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1542..1698] | 3.035979 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)