













| General Information: |
|
| Name(s) found: |
PTC3 /
YBL056W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 468 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
inactivation of MAPK activity involved in osmosensory signaling pathway
[IGI protein amino acid dephosphorylation [IDA regulation of cyclin-dependent protein kinase activity [IMP |
| Molecular Function: |
protein serine/threonine phosphatase activity
[IDA][IEA]
phosphoprotein phosphatase activity [IEA] metal ion binding [IEA] catalytic activity [IEA] hydrolase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGQILSNPII DKEHHSGTDC LTAFGLCAMQ GWRMSMEDAH IVEPNLLAES DEEHLAFYGI 60
61 FDGHGGSSVA EFCGSKMISI LKKQESFKSG MLEQCLIDTF LATDVELLKD EKLKDDHSGC 120
121 TATVILVSQL KKLLICANSG DSRTVLSTGG NSKAMSFDHK PTLLSEKSRI VAADGFVEMD 180
181 RVNGNLALSR AIGDFEFKSN TKLGPHEQVV TCVPDIICHN LNYDEDEFVI LACDGIWDCL 240
241 TSQECVDLVH YGISQGNMTL SDISSRIVDV CCSPTTEGSG IGCDNMSISI VALLKENESE 300
301 SQWFERMRSK NYNIQTSFVQ RRKSIFDFHD FSDDDNEVFA ITTKKLQDRL NRSKDNDDME 360
361 IDDLDTELDS SATPSKLSGE DRTGPIDLFS LEALLEAGIQ IRQRPSSDSD GNTSYFHGAS 420
421 LSDMLASLSN AAAGETEPND ADDNDDNDGE ENGKNENAKK GSKIEEIE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
PAA1 PTC3 |
| View Details | Krogan NJ, et al. (2006) |
|
PAA1 PTC2 PTC3 PTC4 VPS34 |
| View Details | Gavin AC, et al. (2006) |

|
PAA1 PTC3 |
| View Details | Gavin AC, et al. (2002) |
|
ENP1 HRR25 PAA1 PFK1 PTC3 RAD50 TSR1 VPS15 |
| View Details | Ho Y, et al. (2002) |
|
AAC3 AHA1 ATP3 BEM2 COP1 ECM10 ECM29 GCD11 HOM3 HOR2 ILV2 OYE2 PAA1 PIL1 PRB1 PTC3 RAD52 RAD55 RAD59 REP1 RHR2 SEC27 SEC53 TCB1 TDA10 TEF4 UBA1 VMA8 YER138C YHR033W YPT31 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
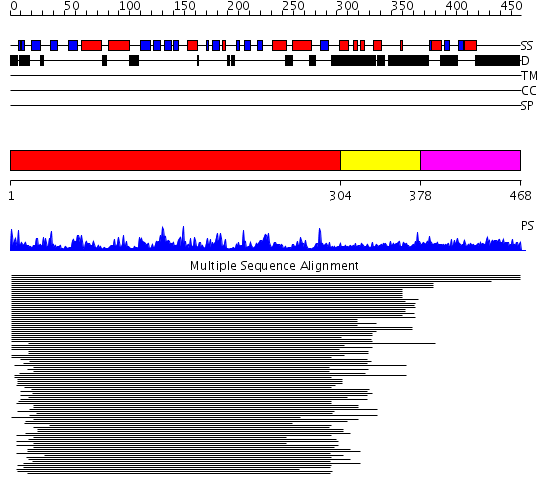
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..303] | 499.85005 | Protein serine/threonine phosphatase 2C, C-terminal domain; Protein serine/threonine phosphatase 2C, catalytic domain | |
| 2 | View Details | [304..377] | 499.85005 | Protein serine/threonine phosphatase 2C, C-terminal domain; Protein serine/threonine phosphatase 2C, catalytic domain | |
| 3 | View Details | [378..468] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)