













| General Information: |
|
| Name(s) found: |
VPS15 /
YBR097W
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1454 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA Golgi membrane [TAS |
| Biological Process: |
protein retention in Golgi apparatus
[IMP telomere maintenance [IMP inositol lipid-mediated signaling [IMP protein amino acid phosphorylation [IDA protein targeting to vacuole [IMP |
| Molecular Function: |
protein serine/threonine kinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGAQLSLVVQ ASPSIAIFSY IDVLEEVHYV SQLNSSRFLK TCKALDPNGE IVIKVFIKPK 60
61 DQYSLRPFLQ RIRAQSFKLG QLPHVLNYSK LIETNRAGYM IRQHLKNNLY DRLSLRPYLQ 120
121 DIELKFIAFQ LLNTLKDIHN LNIVHGDIKT ENILVTSWNW CILTDFAAFI KPVYLPEDNP 180
181 GEFLFYFDTS KRRTCYLAPE RFNSKLYQDG KSNNGRLTKE MDIFSLGCVI AEIFAEGRPI 240
241 FNLSQLFKYK SNSYDVNREF LMEEMNSTDL RNLVLDMIQL DPSKRLSCDE LLNKYRGIFF 300
301 PDYFYTFIYD YFRNLVTMTT STPISDNTCT NSTLEDNVKL LDETTEKIYR DFSQICHCLD 360
361 FPLIKDGGEI GSDPPILESY KIEIEISRFL NTNLYFPQNY HLVLQQFTKV SEKIKSVKEE 420
421 CALLFISYLS HSIRSIVSTA TKLKNLELLA VFAQFVSDEN KIDRVVPYFV CCFEDSDQDV 480
481 QALSLLTLIQ VLTSVRKLNQ LNENIFVDYL LPRLKRLLIS NRQNTNYLRI VFANCLSDLA 540
541 IIINRFQEFT FAQHCNDNSM DNNTEIMESS TKYSAKLIQS VEDLTVSFLT DNDTYVKMAL 600
601 LQNILPLCKF FGRERTNDII LSHLITYLND KDPALRVSLI QTISGISILL GTVTLEQYIL 660
661 PLLIQTITDS EELVVISVLQ SLKSLFKTGL IRKKYYIDIS KTTSPLLLHP NNWIRQFTLM 720
721 IIIEIINKLS KAEVYCILYP IIRPFFEFDV EFNFKSMISC CKQPVSRSVY NLLCSWSVRA 780
781 SKSLFWKKII TNHVDSFGNN RIEFITKNYS SKNYGFNKRD TKSSSSLKGI KTSSTVYSHD 840
841 NKEIPLTAED INWIDKFHII GLTEKDIWKI VALRGYVIRT ARVMAANPDF PYNNSNYRPL 900
901 VQNSPPNLNL TNIMPRNIFF DVEFAEESTS EGQDSNLENQ QIYKYDESEK DSNKLNINGS 960
961 KQLSTVMDIN GSLIFKNKSI ATTTSNLKNV FVQLEPTSYH MHSPNHGLKD NANVKPERKV1020
1021 VVSNSYEGDV ESIEKFLSTF KILPPLRDYK EFGPIQEIVR SPNMGNLRGK LIATLMENEP1080
1081 NSITSSAVSP GETPYLITGS DQGVIKIWNL KEIIVGEVYS SSLTYDCSST VTQITMIPNF1140
1141 DAFAVSSKDG QIIVLKVNHY QQESEVKFLN CECIRKINLK NFGKNEYAVR MRAFVNEEKS1200
1201 LLVALTNLSR VIIFDIRTLE RLQIIENSPR HGAVSSICID EECCVLILGT TRGIIDIWDI1260
1261 RFNVLIRSWS FGDHAPITHV EVCQFYGKNS VIVVGGSSKT FLTIWNFVKG HCQYAFINSD1320
1321 EQPSMEHFLP IEKGLEELNF CGIRSLNALS TISVSNDKIL LTDEATSSIV MFSLNELSSS1380
1381 KAVISPSRFS DVFIPTQVTA NLTMLLRKMK RTSTHSVDDS LYHHDIINSI STCEVDETPL1440
1441 LVACDNSGLI GIFQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
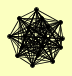
| View Details | Krogan NJ, et al. (2006) |
|
AIM3 BAR1 COA4 GYL1 GYP5 HNT1 KIN2 MEF2 NCP1 PCL10 PRP28 RVS161 RVS167 SAE2 SLK19 TIM21 VPS15 |
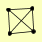
| View Details | Gavin AC, et al. (2006) |
|
VPS15 VPS30 VPS34 VPS38 |
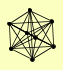
| View Details | Gavin AC, et al. (2002) |
|
ENP1 HRR25 PAA1 PFK1 PTC3 RAD50 TSR1 VPS15 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
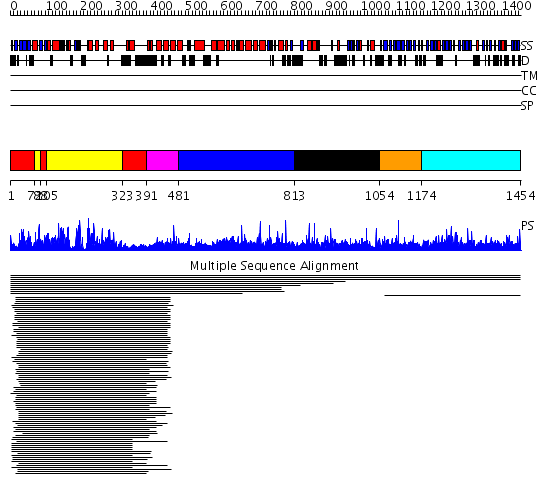
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] [88..104] [323..390] |
174.228787 | cAMP-dependent PK, catalytic subunit | |
| 2 | View Details | [72..87] [105..322] |
174.228787 | cAMP-dependent PK, catalytic subunit | |
| 3 | View Details | [391..480] | 83.39794 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 4 | View Details | [481..812] | 83.39794 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 5 | View Details | [813..1053] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1054..1173] | 4.30103 | beta1-subunit of the signal-transducing G protein heterotrimer | |
| 7 | View Details | [1174..1454] | 14.46 | Tricorn protease; Tricorn protease N-terminal domain; Tricorn protease domain 2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.59 |
Source: Reynolds et al. (2008)