













| General Information: |
|
| Name(s) found: |
HTZ1 /
YOL012C
[SGD]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 134 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromatin assembly complex
[IPI nuclear chromatin [IDA |
| Biological Process: |
chromatin silencing at silent mating-type cassette
[IGI regulation of transcription from RNA polymerase II promoter [IGI |
| Molecular Function: |
chromatin binding
[IDA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
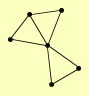
|
ABF2 HTZ1 KAP114 NAP1 RPL17A RPS16B, RPS16A |
| View Details | Krogan NJ, et al. (2006) |
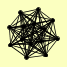
|
GIN4 HFD1 HTZ1 KAP114 KCC4 NAP1 PRO3 REI1 RIM11 RPS6A, RPS6B UBC8 VIP1 WTM1 YKR011C |
| View Details | Gavin AC, et al. (2002) |
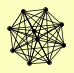
|
GIN4 HTZ1 KAP114 KCC4 LYS12 NAP1 NBA1 NIS1 SSC1 YRA1 |
| View Details | Ho Y, et al. (2002) |
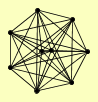
|
GND1 HHF1, HHF2 HTB2 HTZ1 RAD16 RAD7 SHP1 TMA29 TSA2 |
| View Details | Qiu et al. (2008) |
|
ABF1 HHF1, HHF2 HTZ1 PCC1 POB3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
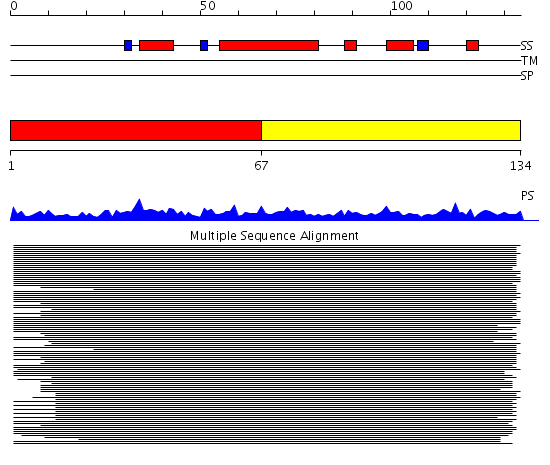
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..66] | 363.201249 | Histone H2A | |
| 2 | View Details | [67..134] | 363.201249 | Histone H2A |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)