













| General Information: |
|
| Name(s) found: |
RKM1 /
YPL208W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 583 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
response to drug
[IMP |
| Molecular Function: |
protein-lysine N-methyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSDALKALL QWGASFGVIV PEELKFLYTD LKGIICVCEK DIDNPSIKIP PEIVISRNLP 60
61 MKFFGLSEST KNINGWLKLF FAKIKFDRDN DTIVDNVRVN DKFKPYLDAL PSRLNSPLVW 120
121 NPSELKRLSS TNIGNSIHEK FEGIFKEWFE LVSSSDMFDL ERVADDVQTF HNLDELTYEA 180
181 LYEKILKITE LQRPTIWYSF PAFLWSHLIF ISRAFPEYVL NRNCPDNSIV LLPIVDLLNH 240
241 DYRSKVKWYP ENGWFCYEKI GTASQSRELS NNYGGKGNEE LLSGYGFVLE DNIFDSVALK 300
301 VKLPLDVVST ILETEPSLKL PLLSDYTTYA FENKDCVQQE KKATRSATDY INGVTYFINI 360
361 QNEQCLEPLL DLFTYLSKAE EEDLHDLRAR LQGIQMLRNA LQSKLNSITG PPATDDSYAI 420
421 DPYRVYCADV YTKGQKQILK EALTRLKKLE KTMLSENKHQ LLTMSKILKN DPAFAETELP 480
481 SLFSNEDGEE VIFESTYDLL ILWILLKTKK NSYPTKYEWV GQQYTNFKQT AYISDDAKAF 540
541 HTAYFEKQDD VDLAEVDHAI QFVVDNSFTR TSSTTEETIL VRK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
BCP1 RKM1 |
| View Details | Krogan NJ, et al. (2006) |
|
RKM1 YMR1 |
| View Details | Gavin AC, et al. (2006) |

|
BCP1 RKM1 |
| View Details | Gavin AC, et al. (2002) |
|
BCP1 GFA1 GLT1 KAP123 NOC2 RKM1 SSA4 YOL098C |
| View Details | Ho Y, et al. (2002) |
|
BCP1 FET4 IMD4 RKM1 RPL23B, RPL23A |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
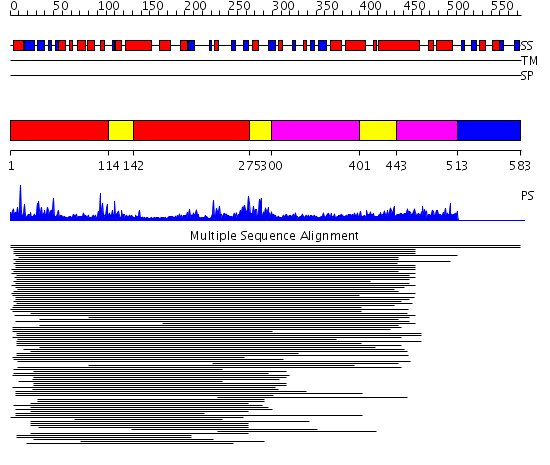
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..113] [142..274] |
72.0 | RuBisCo LSMT C-terminal, substrate-binding domain; RuBisCo LSMT catalytic domain | |
| 2 | View Details | [114..141] [275..299] [401..442] |
72.0 | RuBisCo LSMT C-terminal, substrate-binding domain; RuBisCo LSMT catalytic domain | |
| 3 | View Details | [300..400] [443..512] |
72.0 | RuBisCo LSMT C-terminal, substrate-binding domain; RuBisCo LSMT catalytic domain | |
| 4 | View Details | [513..583] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)