













| General Information: |
|
| Name(s) found: |
YMR1 /
YJR110W
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 688 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
vesicle-mediated transport
[IMP inositol lipid-mediated signaling [IMP phosphoinositide dephosphorylation [IDA |
| Molecular Function: |
phosphatidylinositol-3-phosphatase activity
[IDA phosphatase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEYIKIAKVS NVVLHRRGTA TQGTLHLTTH HLIFESPQLS TEFWFPYPLI YGVHKNPGST 60
61 LLSKLTSTNQ IQLEGTDSQN YKLYQGKDLW SFVNIKVIGK DYAVFSLDFG GDLHLQARKV 120
121 YDSILNLTVL SNITQLYAFI YISNNLERKL PSPDSWDIYD PIKEFRRQGL DSKDETCPWR 180
181 LSTVNEHYEF CPTYPSKLFV PRSTSDILLK HASKFRSQKR IPVLTYHHKA TDCNILRSSQ 240
241 PLPGLINQRS IQDEKLVWES FNSFCNKDIR RTKHVIVDAR PRTNALAQMA LGGGTENMDN 300
301 YNFFLADNNM GVDKSLKLPT VTRLFLGIDN IHIVSNTAAY MTEVICQGGD LNLPLEQNLI 360
361 RSQKFSNWLK LNTLILKSVD MLLKSIIFNH SNVLVHCSDG WDRTSQVVSL LEICLDPFYR 420
421 TFEGFMILVE KDWCSFGHRF LERSGHLNSD IRFHDNTMHS NFNDVDTNGD DLDIGVNTQD 480
481 DYAEDDEGGE DETNLINLSR ISKKFNENFK LNKKSLKFVS PVFQQFLDCV YQLLTQNPDL 540
541 FEFNERFLRR LVYHLYSCQY GTFLSNSEKE KFQQNLPNKT KSVWDYFRSR RKQFINPNFI 600
601 QRKRSGMNEH DQNLEEEEKV EWISPDLKKV QWWWQLYGRK DSEMNDELRH KRDSVPISVD 660
661 KKSKEHSNSD GGKGLNLSIF GFDMFNRK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
RKM1 YMR1 |
| View Details | Ho Y, et al. (2002) |

|
RGR1 YMR1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
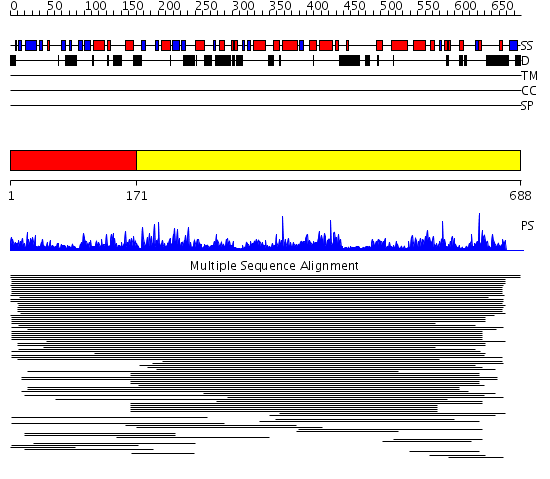
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..170] | 1.045972 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [171..688] | 5.05091 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)