













| General Information: |
|
| Name(s) found: |
ARC15 /
YIL062C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 154 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial envelope
[TAS Arp2/3 protein complex [IDA |
| Biological Process: |
mitochondrion inheritance
[IMP actin cortical patch assembly [IMP |
| Molecular Function: |
actin binding
[TAS structural molecule activity [IMP |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
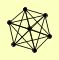
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ARC15 ARC18 ARC19 ARC35 ARC40 ARP2 ARP3 |
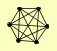
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
ARC15 ARC18 ARC19 ARC35 ARP2 ARP3 |
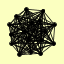
| View Details | Krogan NJ, et al. (2006) |
|
ARC15 ASR1 CAJ1 CDC7 CRN1 INP52 IRC22 MSK1 PNO1 PST2 RAD34 SAC6 SAR1 SLM1 TCP1 TED1 YPP1 |
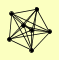
| View Details | Gavin AC, et al. (2006) |
|
ARC15 ARC18 ARC19 ARC35 ARC40 ARP2 ARP3 |
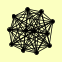
| View Details | Gavin AC, et al. (2002) |
|
ARC15 ARC18 ARC19 ARC35 ARC40 ARP2 ARP3 CCT5 CCT8 NOG2 PFK1 PRT1 |
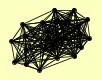
| View Details | Ho Y, et al. (2002) |
|
ADR1 ARC15 ARC18 ARC19 ARC35 ARC40 ARP2 ARP3 ATP3 BNI1 DUR1,2 FET3 GFA1 IMH1 KAP122 MCM4 MCM7 MET18 PST2 PUF3 RPN8 RPT4 RVB1 TRP3 YJR029W |
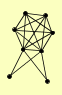
| View Details | Qiu et al. (2008) |
|
ARC15 ARC18 ARC19 ARC35 ARP2 ARP3 ENT1 PFY1 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
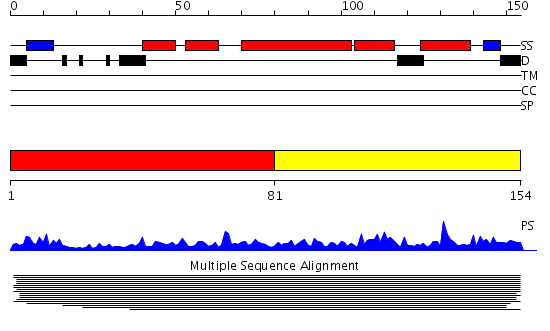
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | 128.9897 | Arp2/3 complex 16 kDa subunit ARPC5 | |
| 2 | View Details | [81..154] | 128.9897 | Arp2/3 complex 16 kDa subunit ARPC5 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)