













| General Information: |
|
| Name(s) found: |
CAJ1 /
YER048C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 391 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
chaperone regulator activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKETEYYDI LGIKPEATPT EIKKAYRRKA METHPDKHPD DPDAQAKFQA VGEAYQVLSD 60
61 PGLRSKYDQF GKEDAVPQQG FEDASEYFTA IFGGDGFKDW IGEFSLFKEL NEATEMFGKE 120
121 DEEGTAATET EKADESTDGG MVKHDTNKAE SLKKDKLSKE QREKLMEMEK KRREDMMKQV 180
181 DELAEKLNEK ISRYLIAVKS NNLEEFTRKL DQEIEDLKLE SFGLELLYLL ARVYKTKANN 240
241 FIMSKKTYGI SKIFTGTRDN ARSVKSAYNL LSTGLEAQKA MEKMSEVNTD ELDQYERAKF 300
301 ESTMAGKALG VMWAMSKFEL ERKLKDVCNK ILNDKKVPSK ERIAKAKAML FIAHKFASAR 360
361 RSPEEAEEAR VFEELILGEQ EKEHKKHTVA R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
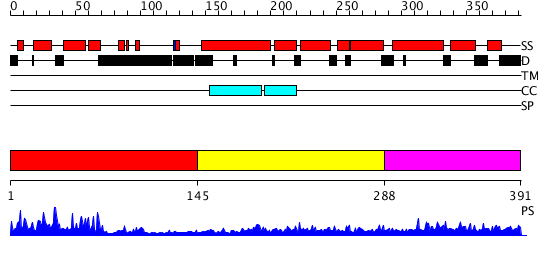
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..144] | 137.69897 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [145..287] | 4.221849 | Heat shock protein 40 Sis1 | |
| 3 | View Details | [288..391] | 1.042991 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)