













| General Information: |
|
| Name(s) found: |
PDC2 /
YDR081C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 925 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC |
| Biological Process: |
transcription from RNA polymerase II promoter
[IMP glucose catabolic process to ethanol [IMP thiamin biosynthetic process [TAS |
| Molecular Function: |
transcription regulator activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSIQQRYNI CLMAERHPKW TQLELAKWAY ETFQLPKIPS QGTISRLLAR KSTYMNCKEH 60
61 EKDANRLRKP NNLLVRKILQ EWISQSLWNG IPITSPIIQD TAQAVWHRIP AEHREGNGSF 120
121 SYKWISNFLS KMDVNISVLD EELPKTPKVW TFEERDVLKA YFSKIPPKDL FTLDEAFLSY 180
181 NLPLDYAQYE ASSIQRRIEV ATVMLCSNLD GSEKLKPVVV GKYDSYKSFR NYFPNEPNDP 240
241 VSQSMLGTKM AKKFDISYHS NRKAWLTSNL FHNWLVRWDK RLVAVNRKIW IVLDDSCCHR 300
301 IINLRLQNIK LVYTSSNSKF LPFNWGVWDE FKTRYRIQQY QALIDLQNRI SKNIQNKNKS 360
361 ERNECIPNGK KCLISFEQSQ LTMSNAFKFI KKAWDDIPVD AIKANWKSSG LLPPEMIHLN 420
421 ENVSMAFKKN EVLESVLNRL CDEYYCVKKW EYEMLLDLNI ENKNTNFLST EELVESAIVE 480
481 PCEPDFDTAP KGNEVHDDNF DVSVFANEDD NNQNHLSMSQ ASHNPDYNSN HSNNAIENTN 540
541 NRGSNNNNNN NGSSNNINDN DSSVKYLQQN TVDNSTKTGN PGQPNISSME SQRNSSTTDL 600
601 VVDGNYDVNF NGLLNDPYNT MKQPGPLDYN VSTLIDKPNL FLSPDLDLST VGVDMQLPSS 660
661 EYFSEVFSSA IRNNEKAASD QNKSTDELPS STAMANSNSI TTALLESRNQ AQPFDVPHMN 720
721 GLLSDTSKSG HSVNSSNAIS QNSLNNFQHN SASVAEASSP SITPSPVAIN STGAPARSII 780
781 SAPIDSNSSA SSPSALEHLE GAVSGMSPSS TTILSNLQTN INIAKSLSTI MKHAESNEIS 840
841 LTKETINELN FNYLTLLKRI KKTRKQLNSE SIKINSKNAQ DHLETLLSGA AAAAATSANN 900
901 LDLPTGGSNL PDSNNLHLPG NTGFF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
GFA1 PDC2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
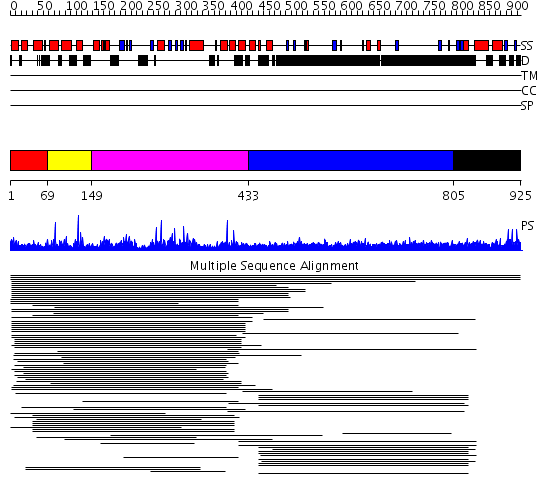
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..68] | 72.69897 | Ars-binding protein 1, ABP1 | |
| 2 | View Details | [69..148] | 72.69897 | Ars-binding protein 1, ABP1 | |
| 3 | View Details | [149..432] | 127.119186 | DDE superfamily endonuclease No confident structure predictions are available. | |
| 4 | View Details | [433..804] | 24.194997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [805..925] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)