GO TERM SUMMARY
|
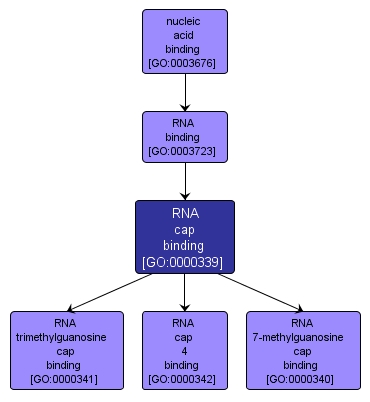
| Name: |
RNA cap binding |
| Acc: |
GO:0000339 |
| Aspect: |
Molecular Function |
| Desc: |
Interacting selectively and non-covalently with a 7-methylguanosine (m7G) moiety or derivative located at the 5' end of an RNA molecule. |
Synonyms:
- binding to mRNA cap
- mRNA cap binding
- snRNA cap binding
|
|

|
INTERACTIVE GO GRAPH
|