













| General Information: |
|
| Name(s) found: |
TRM12 /
YML005W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 462 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
tRNA methylation
[IDA |
| Molecular Function: |
S-adenosylmethionine-dependent methyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDETMPVEF LVSDKRLLKT IKVKLETNGL FVTPIYSDND NKVIKSSIED LNHPLAVEIN 60
61 NIAGVKARFH ESGNLERSEG HLKHQSNSIT EFTKSFLKDH GLANDKIFLS HLLDHLPLKY 120
121 TIYPPVVLFN NSTVRSFNHP IWQKAFQLKL FDPNEYYREL LCFLSPGKPS KGTSLHPNNR 180
181 LLTHLAINNP ITEADVLRRP FNIQPLYGKL IDDSILDDND NTLWENPSQE QLNSSIWCKV 240
241 IQNGVTQIWS PVFTMFSRGN IKEKKRVLTT FPDICNNDVV DLYAGIGYFT FSYLTKGART 300
301 LFAFELNPWS VEGLKRGLKA NGFNKSGNCH VFQESNEMCV QRLTEFLSQN PGFRLRIRHI 360
361 NLGLLPSSKQ GWPLAIKLIY LQGASLEKVT MHIHENVHID AIEDGSFEKN VIVELDAINE 420
421 SIALIRNRGI KLQFVRSKLE RIKTFAPDIW HVCVDVDVIV ST |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
PLB3 RPS10B SCM3 SUP35 SUP45 TRM12 VPS3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
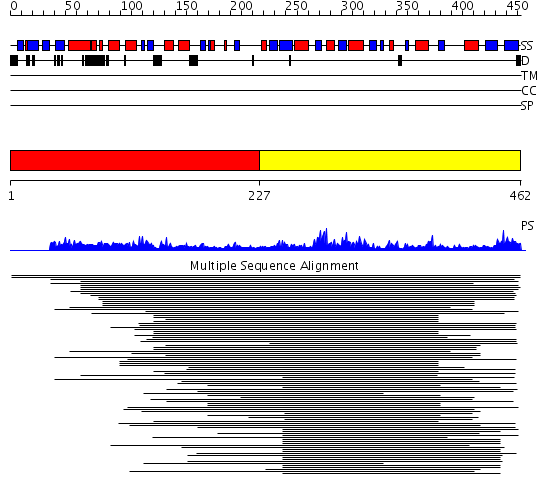
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..226] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [227..462] | 26.221849 | Arginine methyltransferase, HMT1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)