













| General Information: |
|
| Name(s) found: |
LYS2 /
YBR115C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1392 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
lysine biosynthetic process via aminoadipic acid
[IDA |
| Molecular Function: |
L-aminoadipate-semialdehyde dehydrogenase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTNEKVWIEK LDNPTLSVLP HDFLRPQQEP YTKQATYSLQ LPQLDVPHDS FSNKYAVALS 60
61 VWAALIYRVT GDDDIVLYIA NNKILRFNIQ PTWSFNELYS TINNELNKLN SIEANFSFDE 120
121 LAEKIQSCQD LERTPQLFRL AFLENQDFKL DEFKHHLVDF ALNLDTSNNA HVLNLIYNSL 180
181 LYSNERVTIV ADQFTQYLTA ALSDPSNCIT KISLITASSK DSLPDPTKNL GWCDFVGCIH 240
241 DIFQDNAEAF PERTCVVETP TLNSDKSRSF TYRDINRTSN IVAHYLIKTG IKRGDVVMIY 300
301 SSRGVDLMVC VMGVLKAGAT FSVIDPAYPP ARQTIYLGVA KPRGLIVIRA AGQLDQLVED 360
361 YINDELEIVS RINSIAIQEN GTIEGGKLDN GEDVLAPYDH YKDTRTGVVV GPDSNPTLSF 420
421 TSGSEGIPKG VLGRHFSLAY YFNWMSKRFN LTENDKFTML SGIAHDPIQR DMFTPLFLGA 480
481 QLYVPTQDDI GTPGRLAEWM SKYGCTVTHL TPAMGQLLTA QATTPFPKLH HAFFVGDILT 540
541 KRDCLRLQTL AENCRIVNMY GTTETQRAVS YFEVKSKNDD PNFLKKLKDV MPAGKGMLNV 600
601 QLLVVNRNDR TQICGIGEIG EIYVRAGGLA EGYRGLPELN KEKFVNNWFV EKDHWNYLDK 660
661 DNGEPWRQFW LGPRDRLYRT GDLGRYLPNG DCECCGRADD QVKIRGFRIE LGEIDTHISQ 720
721 HPLVRENITL VRKNADNEPT LITFMVPRFD KPDDLSKFQS DVPKEVETDP IVKGLIGYHL 780
781 LSKDIRTFLK KRLASYAMPS LIVVMDKLPL NPNGKVDKPK LQFPTPKQLN LVAENTVSET 840
841 DDSQFTNVER EVRDLWLSIL PTKPASVSPD DSFFDLGGHS ILATKMIFTL KKKLQVDLPL 900
901 GTIFKYPTIK AFAAEIDRIK SSGGSSQGEV VENVTANYAE DAKKLVETLP SSYPSREYFV 960
961 EPNSAEGKTT INVFVTGVTG FLGSYILADL LGRSPKNYSF KVFAHVRAKD EEAAFARLQK1020
1021 AGITYGTWNE KFASNIKVVL GDLSKSQFGL SDEKWMDLAN TVDIIIHNGA LVHWVYPYAK1080
1081 LRDPNVISTI NVMSLAAVGK PKFFDFVSST STLDTEYYFN LSDKLVSEGK PGILESDDLM1140
1141 NSASGLTGGY GQSKWAAEYI IRRAGERGLR GCIVRPGYVT GASANGSSNT DDFLLRFLKG1200
1201 SVQLGKIPDI ENSVNMVPVD HVARVVVATS LNPPKENELA VAQVTGHPRI LFKDYLYTLH1260
1261 DYGYDVEIES YSKWKKSLEA SVIDRNEENA LYPLLHMVLD NLPESTKAPE LDDRNAVASL1320
1321 KKDTAWTGVD WSNGIGVTPE EVGIYIAFLN KVGFLPPPTH NDKLPLPSIE LTQAQISLVA1380
1381 SGAGARGSSA AA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
LYS2 LYS5 |
| View Details | Krogan NJ, et al. (2006) |
|
FES1 LSG1 LYS2 |
| View Details | Gavin AC, et al. (2006) |
|
IMD2 LYS2 LYS5 SXM1 |
| View Details | Gavin AC, et al. (2002) |
|
ADH1 ECM1 GCN1 HSC82 KAP123 LYS2 LYS5 PSE1 SXM1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
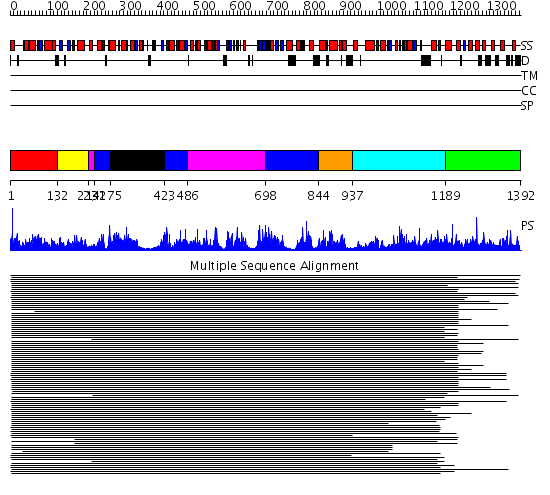
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..131] | 1.206982 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [132..213] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [214..230] [486..697] |
635.0103 | Phenylalanine activating domain of gramicidin synthetase 1 | |
| 4 | View Details | [231..274] [423..485] [698..843] |
635.0103 | Phenylalanine activating domain of gramicidin synthetase 1 | |
| 5 | View Details | [275..422] | 635.0103 | Phenylalanine activating domain of gramicidin synthetase 1 | |
| 6 | View Details | [844..936] | 70.440336 | Peptidyl carrier protein (PCP), thioester domain | |
| 7 | View Details | [937..1188] | 6.045757 | VibH | |
| 8 | View Details | [1189..1392] | 1.087943 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)