













| General Information: |
|
| Name(s) found: |
RIF1 /
YBR275C
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1916 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear telomere cap complex
[IDA |
| Biological Process: |
telomere maintenance
[IMP telomere maintenance via telomerase [IMP chromatin silencing at telomere [IMP |
| Molecular Function: |
protein binding
[IGI telomeric DNA binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKDFSDKKK HTIDRIDQHI LRRSQHDNYS NGSSPWMKTN LPPPSPQAHM HIQSDLSPTP 60
61 KRRKLASSSD CENKQFDLSA INKNLYPEDT GSRLMQSLPE LSASNSDNVS PVTKSVAFSD 120
121 RIESSPIYRI PGSSPKPSPS SKPGKSILRN RLPSVRTVSD LSYNKLQYTQ HKLHNGNIFT 180
181 SPYKETRVNP RALEYWVSGE IHGLVDNESV SEFKEIIEGG LGILRQESED YVARRFEVYA 240
241 TFNNIIPILT TKNVNEVDQK FNILIVNIES IIEICIPHLQ IAQDTLLSSS EKKNPFVIRL 300
301 YVQIVRFFSA IMSNFKIVKW LTKRPDLVNK LKVIYRWTTG ALRNENSNKI IITAQVSFLR 360
361 DEKFGTFFLS NEEIKPIIST FTEIMEINSH NLIYEKLLLI RGFLSKYPKL MIETVTSWLP 420
421 GEVLPRIIIG DEIYSMKILI TSIVVLLELL KKCLDFVDEH ERIYQCIMLS PVCETIPEKF 480
481 LSKLPLNSYD SANLDKVTIG HLLTQQIKNY IVVKNDNKIA MDLWLSMTGL LYDSGKRVYD 540
541 LTSESNKVWF DLNNLCFINN HPKTRLMSIK VWRIITYCIC TKISQKNQEG NKSLLSLLRT 600
601 PFQMTLPYVN DPSAREGIIY HLLGVVYTAF TSNKNLSTDM FELFWDHLIT PIYEDYVFKY 660
661 DSIHLQNVLF TVLHLLIGGK NADVALERKY KKHIHPMSVI ASEGVKLKDI SSLPPQIIKR 720
721 EYDKIMKVVF QTVEVAISNV NLAHDLILTS LKHLPEDRKD QTHLESFSSL ILKVTQNNKD 780
781 TPIFRDFFGA VTSSFVYTFL DLFLRKNDSS LVNFNIQISK VGISQGNMTL DLLKDVIRKA 840
841 RNETSEFLII EKFLELDDKK TEVYAQNWVG STLLPPNISF REFQSLANIV NKVPNENSIE 900
901 NFLDLCLKLS FPVNLFTLLH VSMWSNNNFI YFIQSYVSKN ENKLNVDLIT LLKTSLPGNP 960
961 ELFSGLLPFL RRNKFMDILE YCIHSNPNLL NSIPDLNSDL LLKLLPRSRA SYFAANIKLF1020
1021 KCSEQLTLVR WLLKGQQLEQ LNQNFSEIEN VLQNASDSEL EKSEIIRELL HLAMANPIEP1080
1081 LFSGLLNFCI KNNMADHLDE FCGNMTSEVL FKISPELLLK LLTYKEKPNG KLLAAVIEKI1140
1141 ENGDDDYILE LLEKIIIQKE IQILEKLKEP LLVFFLNPVS SNMQKHKKST NMLRELVLLY1200
1201 LTKPLSRSAA KKFFSMLISI LPPNPNYQTI DMVNLLIDLI KSHNRKFKDK RTYNATLKTI1260
1261 GKWIQESGVV HQGDSSKEIE AIPDTKSMYI PCEGSENKLS NLQRKVDSQD IQVPATQGMK1320
1321 EPPSSIQISS QISAKDSDSI SLKNTAIMNS SQQESHANRS RSIDDETLEE VDNESIREID1380
1381 QQMKSTQLDK NVANHSNICS TKSDEVDVTE LHESIDTQSS EVNAYQPIEV LTSELKAVTN1440
1441 RSIKTNPDHN VVNSDNPLKR PSKETPTSEN KRSKGHETMV DVLVSEEQAV SPSSDVICTN1500
1501 IKSIANEESS LALRNSIKVE TNCNENSLNV TLDLDQQTIT KEDGKGQVEH VQRQENQESM1560
1561 NKINSKSFTQ DNIAQYKSVK KARPNNEGEN NDYACNVEQA SPVRNEVPGD GIQIPSGTIL1620
1621 LNSSKQTEKS KVDDLRSDED EHGTVAQEKH QVGAINSRNK NNDRMDSTPI QGTEEESREV1680
1681 VMTEEGINVR LEDSGTCELN KNLKGPLKGD KDANINDDFV PVEENVRDEG FLKSMEHAVS1740
1741 KETGLEEQPE VADISVLPEI RIPIFNSLKM QGSKSQIKEK LKKRLQRNEL MPPDSPPRMT1800
1801 ENTNINAQNG LDTVPKTIGG KEKHHEIQLG QAHTEADGEP LLGGDGNEDA TSREATPSLK1860
1861 VHFFSKKSRR LVARLRGFTP GDLNGISVEE RRNLRIELLD FMMRLEYYSN RDNDMN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
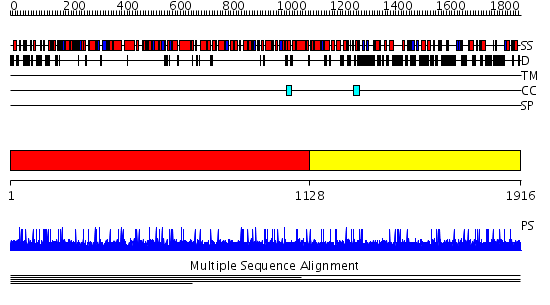
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1127] | 1.003001 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [1128..1916] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [291..486] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [487..712] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [713..779] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [780..876] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [877..1110] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1111..1248] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1249..1491] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1492..1669] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1670..1916] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)