













| General Information: |
|
| Name(s) found: |
PGI1 /
YBR196C
[SGD]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 554 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA cytosol [TAS] |
| Biological Process: |
gluconeogenesis
[IMP pentose-phosphate shunt [IMP glycolysis [IMP |
| Molecular Function: |
glucose-6-phosphate isomerase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNNSFTNFK LATELPAWSK LQKIYESQGK TLSVKQEFQK DAKRFEKLNK TFTNYDGSKI 60
61 LFDYSKNLVN DEIIAALIEL AKEANVTGLR DAMFKGEHIN STEDRAVYHV ALRNRANKPM 120
121 YVDGVNVAPE VDSVLKHMKE FSEQVRSGEW KGYTGKKITD VVNIGIGGSD LGPVMVTEAL 180
181 KHYAGVLDVH FVSNIDGTHI AETLKVVDPE TTLFLIASKT FTTAETITNA NTAKNWFLSK 240
241 TGNDPSHIAK HFAALSTNET EVAKFGIDTK NMFGFESWVG GRYSVWSAIG LSVALYIGYD 300
301 NFEAFLKGAE AVDNHFTQTP LEDNIPLLGG LLSVWYNNFF GAQTHLVAPF DQYLHRFPAY 360
361 LQQLSMESNG KSVTRGNVFT DYSTGSILFG EPATNAQHSF FQLVHQGTKL IPSDFILAAQ 420
421 SHNPIENKLH QKMLASNFFA QAEALMVGKD EEQVKAEGAT GGLVPHKVFS GNRPTTSILA 480
481 QKITPATLGA LIAYYEHVTF TEGAIWNINS FDQWGVELGK VLAKVIGKEL DNSSTISTHD 540
541 ASTNGLINQF KEWM |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ADH1 HSC82 PGI1 |
| View Details | Krogan NJ, et al. (2006) |
|
AIM22 DPH1 DPH2 EFT2, EFT1 HGH1 MNN1 PGI1 RRG10 TRX1 YMR315W |
| View Details | Gavin AC, et al. (2006) |
|
GAA1 PGI1 SEC1 |
| View Details | Gavin AC, et al. (2002) |
|
ADH1 GIN4 HMO1 KCC4 PBP2 PDC1 PGI1 RAD1 RAD10 SEC1 YEF3 YRA1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Widlund PO, et al. (2005) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
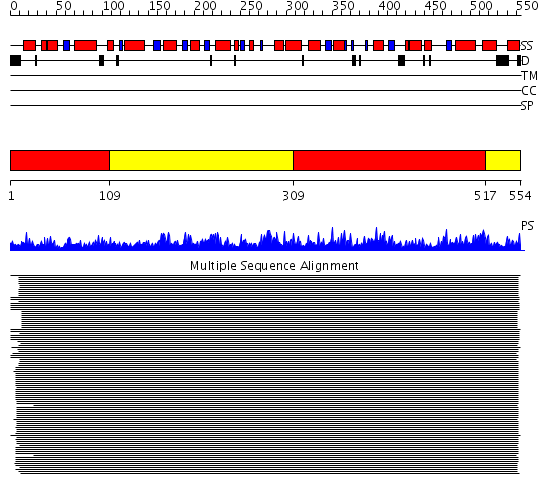
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..108] [309..516] |
11000.0 | Phosphoglucose isomerase, PGI | |
| 2 | View Details | [109..308] [517..554] |
11000.0 | Phosphoglucose isomerase, PGI |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)