













| General Information: |
|
| Name(s) found: |
RSM22 /
YKL155C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 628 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial small ribosomal subunit [IDA |
| Biological Process: |
translation
[IDA |
| Molecular Function: |
structural constituent of ribosome
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMKRCFSILP QNVRFSSKFT SLNLPKLDLA DFIDSNKRGI NVLPSYRDET ASTTQATNSK 60
61 ELRLLSKTLQ GQSYRDQLEL NPDVSKAINN NIMAVHIPNN LRRVATNYYK EIQEPNSLHR 120
121 PCRTKMEVDA HIASIFLQNY GSIFQSLKEL QKRVGPDNFK PQRILDVGYG PATGIVALND 180
181 ILGPNYRPDL KDAVILGNAE MQERAKIILS RQLNEVVDTV EENVSTEKEQ ETDRRNKNFQ 240
241 EDEHIGEVMT KKINIMTNLR SSIPASKEYD LIILTHQLLH DGNQFPIQVD ENIEHYLNIL 300
301 APGGHIVIIE RGNPMGFEII ARARQITLRP ENFPDEFGKI PRPWSRGVTV RGKKDAELGN 360
361 ISSNYFLKVI APCPHQRKCP LQVGNPNFYT HKEGKDLKFC NFQKSIKRPK FSIELKKGKL 420
421 LATSWDGSQG NASRLKGTGR RNGRDYEILN YSYLIFERSH KDENTLKEIK KLRNENVNGK 480
481 YDIGSLGDDT QNSWPRIIND PVKRKGHVMM DLCAPSGELE KWTVSRSFSK QIYHDARKSK 540
541 KGDLWASAAK TQIKGLGDLN VKKFHKLEKE RIKQLKKEER QKARKAMESY NELEDSLQFD 600
601 DHQFSNFEVM KKLSTFHGND FLQHVNRK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
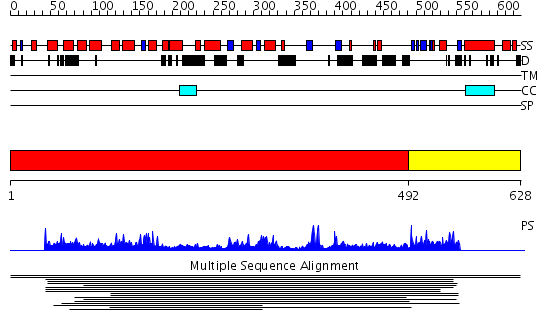
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..491] | 3.011936 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [492..628] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [396..548] | 2.063984 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [549..628] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)