













| General Information: |
|
| Name(s) found: |
ENO1 /
YGR254W
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 437 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA phosphopyruvate hydratase complex [IDA vacuole [IDA |
| Biological Process: |
gluconeogenesis
[IEP regulation of vacuole fusion, non-autophagic [IDA glycolysis [IMP |
| Molecular Function: |
phosphopyruvate hydratase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAVSKVYARS VYDSRGNPTV EVELTTEKGV FRSIVPSGAS TGVHEALEMR DGDKSKWMGK 60
61 GVLHAVKNVN DVIAPAFVKA NIDVKDQKAV DDFLISLDGT ANKSKLGANA ILGVSLAASR 120
121 AAAAEKNVPL YKHLADLSKS KTSPYVLPVP FLNVLNGGSH AGGALALQEF MIAPTGAKTF 180
181 AEALRIGSEV YHNLKSLTKK RYGASAGNVG DEGGVAPNIQ TAEEALDLIV DAIKAAGHDG 240
241 KIKIGLDCAS SEFFKDGKYD LDFKNPNSDK SKWLTGPQLA DLYHSLMKRY PIVSIEDPFA 300
301 EDDWEAWSHF FKTAGIQIVA DDLTVTNPKR IATAIEKKAA DALLLKVNQI GTLSESIKAA 360
361 QDSFAAGWGV MVSHRSGETE DTFIADLVVG LRTGQIKTGA PARSERLAKL NQLLRIEEEL 420
421 GDNAVFAGEN FHHGDKL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ADH1 EFT2, EFT1 ENO1 FBA1 PDC1 |
| View Details | Gavin AC, et al. (2006) |

|
ENO1 NAT1 |
| View Details | Gavin AC, et al. (2002) |
|
ARD1 ASC1 ENO1 MIS1 MYO1 NAT1 PKC1 SSC1 UTP22 |
| View Details | Qiu et al. (2008) |

|
ENO1 ENO2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | #20 Asynchronous Prep1-TiO2 Phosphopeptide enrichment, PartB | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #22 Asynchronous Prep2-IMAC Phosphopeptide enrichment B1-2 | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #05 Alpha Factor Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #07 Alpha Factor Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
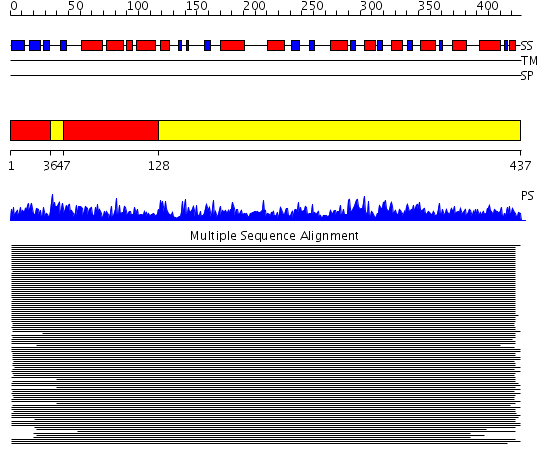
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..35] [47..127] |
11000.0 | Enolase | |
| 2 | View Details | [36..46] [128..437] |
11000.0 | Enolase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)