













| General Information: |
|
| Name(s) found: |
NAT1 /
YDL040C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 854 amino acids |
Gene Ontology: |
|
| Cellular Component: |
NatA complex
[IDA mitochondrion [IDA |
| Biological Process: |
N-terminal protein amino acid acetylation
[IDA |
| Molecular Function: |
peptide alpha-N-acetyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRKRSTKPK PAAKIALKKE NDQFLEALKL YEGKQYKKSL KLLDAILKKD GSHVDSLALK 60
61 GLDLYSVGEK DDAASYVANA IRKIEGASAS PICCHVLGIY MRNTKEYKES IKWFTAALNN 120
121 GSTNKQIYRD LATLQSQIGD FKNALVSRKK YWEAFLGYRA NWTSLAVAQD VNGERQQAIN 180
181 TLSQFEKLAE GKISDSEKYE HSECLMYKND IMYKAASDNQ DKLQNVLKHL NDIEPCVFDK 240
241 FGLLERKATI YMKLGQLKDA SIVYRTLIKR NPDNFKYYKL LEVSLGIQGD NKLKKALYGK 300
301 LEQFYPRCEP PKFIPLTFLQ DKEELSKKLR EYVLPQLERG VPATFSNVKP LYQRRKSKVS 360
361 PLLEKIVLDY LSGLDPTQDP IPFIWTNYYL SQHFLFLKDF PKAQEYIDAA LDHTPTLVEF 420
421 YILKARILKH LGLMDTAAGI LEEGRQLDLQ DRFINCKTVK YFLRANNIDK AVEVASLFTK 480
481 NDDSVNGIKD LHLVEASWFI VEQAEAYYRL YLDRKKKLDD LASLKKEVES DKSEQIANDI 540
541 KENQWLVRKY KGLALKRFNA IPKFYKQFED DQLDFHSYCM RKGTPRAYLE MLEWGKALYT 600
601 KPMYVRAMKE ASKLYFQMHD DRLKRKSDSL DENSDEIQNN GQNSSSQKKK AKKEAAAMNK 660
661 RKETEAKSVA AYPSDQDNDV FGEKLIETST PMEDFATEFY NNYSMQVRED ERDYILDFEF 720
721 NYRIGKLALC FASLNKFAKR FGTTSGLFGS MAIVLLHATR NDTPFDPILK KVVTKSLEKE 780
781 YSENFPLNEI SNNSFDWLNF YQEKFGKNDI NGLLFLYRYR DDVPIGSSNL KEMIISSLSP 840
841 LEPHSQNEIL QYYL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
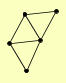
|
ARD1 NAT1 NAT5 RPS8B, RPS8A UTP22 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
ARD1 NAT1 |
| View Details | Krogan NJ, et al. (2006) |
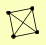
|
ARD1 NAT1 NAT5 RIM8 |
| View Details | Gavin AC, et al. (2006) |
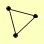
|
ASC1 ENO1 NAT1 |
| View Details | Gavin AC, et al. (2002) |
|
ARD1 ASC1 ENO1 FKS1 GCD6 MIS1 MYO1 NAT1 NEM1 SWI3 UTP22 |
| View Details | Ho Y, et al. (2002) |
|
AAC3 ATP3 DUN1 GPH1 HOR2 NAT1 PFK1 RNQ1 SML1 |
| View Details | Qiu et al. (2008) |
|
ARD1 MAK10 MAK3 MAK31 MDM20 NAT1 NAT3 NAT5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
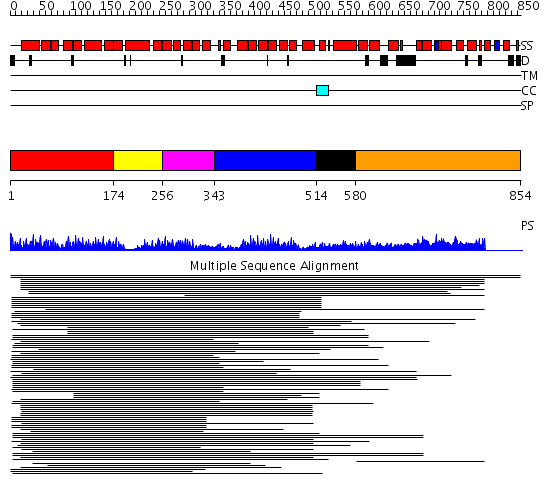
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..173] | 28.0 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 2 | View Details | [174..255] | 28.0 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 3 | View Details | [256..342] | 28.0 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 4 | View Details | [343..513] | 20.957997 | View MSA. Confident ab initio structure predictions are available. | |
| 5 | View Details | [514..579] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [580..854] | 1.183954 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)