













| General Information: |
|
| Name(s) found: |
MLH3 /
YPL164C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 715 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IPI |
| Biological Process: |
reciprocal meiotic recombination
[TAS mismatch repair [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQHIRKLDS DVSERLKSQA CTVSLASAVR EIVQNSVDAH ATTIDVMIDL PNLSFAVYDD 60
61 GIGLTRSDLN ILATQNYTSK IRKMNDLVTM KTYGYRGDAL YSISNVSNLF VCSKKKDYNS 120
121 AWMRKFPSKS VMLSENTILP IDPFWKICPW SRTKSGTVVI VEDMLYNLPV RRRILKEEPP 180
181 FKTFNTIKAD MLQILVMHPM ISLNVQYTDK LRINTEVLFR SKNITEGLTK HQQMSQVLRN 240
241 VFGAIIPPDM LKKVSLKFNE YQIEGIISKM PVGLKDLQFI YINGRRYADS AFQGYVDSLF 300
301 QAQDFGEKGM SLLKTKSVGK PYRSHPVFIL DVRCPQTIDD LLQDPAKKIV KPSHIRTIEP 360
361 LIVKTIRSFL TFQGYLTPDK SDSSFEIVNC SQKTATLPDS RIQISKRNQV LNSKMKIARI 420
421 NSYIGKPAVN GCRINNSTIN YEKIKNIRID GQKSRLRNKL SSRPYDSGFT EDYDSIGKTI 480
481 TDFSISRSVL AKYEVINQVD KKFILIRCLD QSIHNCPLLV LVDQHACDER IRLEELFYSL 540
541 LTEVVTGTFV ARDLKDCCIE VDRTEADLFK HYQSEFKKWG IGYETIEGTM ETSLLEIKTL 600
601 PEMLTSKYNG DKDYLKMVLL QHAHDLKDFK KLPMDLSHFE NYTSVDKLYW WKYSSCVPTV 660
661 FHEILNSKAC RSAVMFGDEL TRQECIILIS KLSRCHNPFE CAHGRPSMVP IAELK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
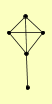
|
ILV2 MLH3 MLP2 QCR2 TEP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data

Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..376] | 100.0 | DNA mismatch repair protein MutL | |
| 2 | View Details | [377..492] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [493..654] | 19.221849 | Crystal structure of the MutL C-terminal domain | |
| 4 | View Details | [655..715] | 2.02 | Crystal structure of the MutL C-terminal domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)