













| General Information: |
|
| Name(s) found: |
CYS4 /
YGR155W
[SGD]
|
| Description(s) found:
Found 34 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 507 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA |
| Biological Process: |
response to drug
[IMP cysteine biosynthetic process [IMP |
| Molecular Function: |
cystathionine beta-synthase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTKSEQQADS RHNVIDLVGN TPLIALKKLP KALGIKPQIY AKLELYNPGG SIKDRIAKSM 60
61 VEEAEASGRI HPSRSTLIEP TSGNTGIGLA LIGAIKGYRT IITLPEKMSN EKVSVLKALG 120
121 AEIIRTPTAA AWDSPESHIG VAKKLEKEIP GAVILDQYNN MMNPEAHYFG TGREIQRQLE 180
181 DLNLFDNLRA VVAGAGTGGT ISGISKYLKE QNDKIQIVGA DPFGSILAQP ENLNKTDITD 240
241 YKVEGIGYDF VPQVLDRKLI DVWYKTDDKP SFKYARQLIS NEGVLVGGSS GSAFTAVVKY 300
301 CEDHPELTED DVIVAIFPDS IRSYLTKFVD DEWLKKNNLW DDDVLARFDS SKLEASTTKY 360
361 ADVFGNATVK DLHLKPVVSV KETAKVTDVI KILKDNGFDQ LPVLTEDGKL SGLVTLSELL 420
421 RKLSINNSNN DNTIKGKYLD FKKLNNFNDV SSYNENKSGK KKFIKFDENS KLSDLNRFFE 480
481 KNSSAVITDG LKPIHIVTKM DLLSYLA |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Dam1 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Dam1 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
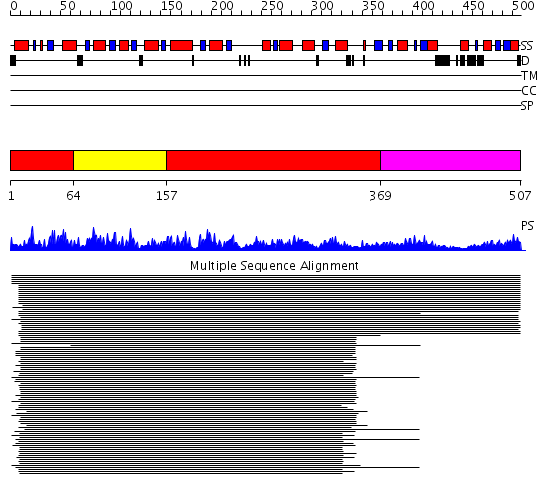
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..63] [157..368] |
1095.9897 | Cystathionine beta-synthase | |
| 2 | View Details | [64..156] | 1095.9897 | Cystathionine beta-synthase | |
| 3 | View Details | [369..507] | 14.2 | Inosine monophosphate dehydrogenase (IMPDH) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)