













| General Information: |
|
| Name(s) found: |
PUF6 /
YDR496C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 656 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nucleolus [IDA |
| Biological Process: |
regulation of transcription, mating-type specific
[IDA ribosome biogenesis [RCA |
| Molecular Function: |
mRNA binding
[IDA specific transcriptional repressor activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPLTKKTNG KRSAKEVSHS EKKLAKKPRI SIDSSDEESE LSKKEDAVSS SSDDDDLDDL 60
61 STSDSEAEEE ADELDISDDS EEHENENEEK EGKDKSEGGE NGNHTEQRKL LKERKMQRKS 120
121 GTQVQQIKSV WERLRVKTPP LPKQIREKLS NEIWELSKDC ISDLVLKHDA SRIVQTLVKY 180
181 SSKDRREQIV DALKGKFYVL ATSAYGKYLL VKLLHYGSRS SRQTIINELH GSLRKLMRHR 240
241 EGAYVVEDLF VLYATHEQRQ QMIKEFWGSE YAVFRETHKD LTIEKVCESS IEKRNIIARN 300
301 LIGTITASVE KGSTGFQILH AAMREYVKIA NEKEISEMIE LLHEQFAELV HTPEGSDVAC 360
361 TLVARANAKE RKLILKALKN HAEKLIKNEY GNIVFITILN CVDDTVLVFK TFSPTVKEHL 420
421 QEFIIDKFGR RPWLYILLGL DGKYFSPIVK NELLRYIELS KATSKKDPLQ RRHELLSKFA 480
481 PMFLSTISKD YSSILTENLG CQFIAEVLIN DELYAQLNEK DQEKYQQVLN NILTTFKGDI 540
541 TEEEHPIHRA FSTRLLKALI QGGKWNNKEK KVIPLKNVQG LGVPFAEKLY DEIIDSSNLL 600
601 EWINNADSSF TIVALYETLK DQKEGKPFLK DLRGVQSKIT TDESNKGSQL LAKLLK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
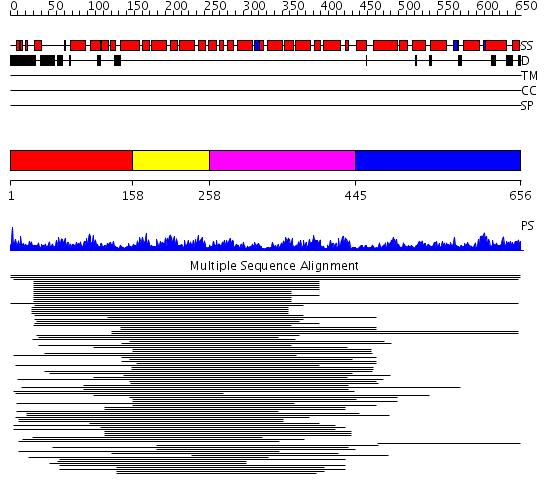
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..157] | 131.67837 | Pumilio 1 | |
| 2 | View Details | [158..257] | 131.67837 | Pumilio 1 | |
| 3 | View Details | [258..444] | 131.67837 | Pumilio 1 | |
| 4 | View Details | [445..656] | 1.032956 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)