













| General Information: |
|
| Name(s) found: |
UTP20 /
YBL004W
[SGD]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2493 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA nuclear outer membrane [IMP nucleolus [IDA small nucleolar ribonucleoprotein complex [IPI |
| Biological Process: |
endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)
[IMP]
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) [IMP] ribosome biogenesis [IEA] endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) [IMP] rRNA processing [IEA][RCA] |
| Molecular Function: |
snoRNA binding
[RCA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKQRQTTKS SKRYRYSSFK ARIDDLKIEP ARNLEKRVHD YVESSHFLAS FDQWKEINLS 60
61 AKFTEFAAEI EHDVQTLPQI LYHDKKIFNS LVSFINFHDE FSLQPLLDLL AQFCHDLGPD 120
121 FLKFYEEAIK TLINLLDAAI EFESSNVFEW GFNCLAYIFK YLSKFLVKKL VLTCDLLIPL 180
181 LSHSKEYLSR FSAEALSFLV RKCPVSNLRE FVRSVFEKLE GDDEQTNLYE GLLILFTESM 240
241 TSTQETLHSK AKAIMSVLLH EALTKSSPER SVSLLSDIWM NISKYASIES LLPVYEVMYQ 300
301 DFNDSLDATN IDRILKVLTT IVFSESGRKI PDWNKITILI ERIMSQSENC ASLSQDKVAF 360
361 LFALFIRNSD VKTLTLFHQK LFNYALTNIS DCFLEFFQFA LRLSYERVFS FNGLKFLQLF 420
421 LKKNWQSQGK KIALFFLEVD DKPELQKVRE VNFPEEFILS IRDFFVTAEI NDSNDLFEIY 480
481 WRAIIFKYSK LQNTEIIIPL LERIFSTFAS PDNFTKDMVG TLLKIYRKED DASGNNLLKT 540
541 ILDNYENYKE SLNFLRGWNK LVSNLHPSES LKGLMSHYPS LLLSLTDNFM LPDGKIRYET 600
601 LELMKTLMIL QGMQVPDLLS SCMVIEEIPL TLQNARDLTI RIKNVGAEFG KTKTDKLVSS 660
661 FFLKYLFGLL TVRFSPVWTG VFDTLPNVYT KDEALVWKLV LSFIKLPDEN QNLDYYQPLL 720
721 EDGANKVLWD SSVVRLRDTI DTFSHIWSKY STQNTSIIST TIERRGNTTY PILIRNQALK 780
781 VMLSIPQVAE NHFVDIAPFV YNDFKTYKDE EDMENERVIT GSWTEVDRNV FLKTLSKFKN 840
841 IKNVYSATEL HDHLMVLLGS RNTDVQKLAL DALLAYKNPT LNKYRDNLKN LLDDTLFKDE 900
901 ITTFLTENGS QSIKAEDEKV VMPYVLRIFF GRAQVPPTSG QKRSRKIAVI SVLPNFKKPY 960
961 INDFLSLASE RLDYNYFFGN SHQINSSKAT LKTIRRMTGF VNIVNSTLSV LRTNFPLHTN1020
1021 SVLQPLIYSI AMAYYVLDTE STEEVHLRKM ASNLRQQGLK CLSSVFEFVG NTFDWSTSME1080
1081 DIYAVVVKPR ISHFSDENLQ QPSSLLRLFL YWAHNPSLYQ FLYYDEFATA TALMDTISNQ1140
1141 HVKEAVIGPI IEAADSIIRN PVNDDHYVDL VTLICTSCLK ILPSLYVKLS DSNSISTFLN1200
1201 LLVSITEMGF IQDDHVRSRL ISSLISILKG KLKKLQENDT QKILKILKLI VFNYNCSWSD1260
1261 IEELYTTISS LFKTFDERNL RVSLTELFIE LGRKVPELES ISKLVADLNS YSSSRMHEYD1320
1321 FPRILSTFKG LIEDGYKSYS ELEWLPLLFT FLHFINNKEE LALRTNASHA IMKFIDFINE1380
1381 KPNLNEASKS ISMLKDILLP NIRIGLRDSL EEVQSEYVSV LSYMVKNTKY FTDFEDMAIL1440
1441 LYNGDEEADF FTNVNHIQLH RRQRAIKRLG EHAHQLKDNS ISHYLIPMIE HYVFSDDERY1500
1501 RNIGNETQIA IGGLAQHMSW NQYKALLRRY ISMLKTKPNQ MKQAVQLIVQ LSVPLRETLR1560
1561 IVRDGAESKL TLSKFPSNLD EPSNFIKQEL YPTLSKILGT RDDETIIERM PIAEALVNIV1620
1621 LGLTNDDITN FLPSILTNIC QVLRSKSEEL RDAVRVTLGK ISIILGAEYL VFVIKELMAT1680
1681 LKRGSQIHVL SYTVHYILKS MHGVLKHSDL DTSSSMIVKI IMENIFGFAG EEKDSENYHT1740
1741 KVKEIKSNKS YDAGEILASN ISLTEFGTLL SPVKALLMVR INLRNQNKLS ELLRRYLLGL1800
1801 NHNSDSESES ILKFCHQLFQ ESEMSNSPQI PKKKVKDQVD EKEDFFLVNL ESKSYTINSN1860
1861 SLLLNSTLQK FALDLLRNVI TRHRSFLTVS HLEGFIPFLR DSLLSENEGV VISTLRILIT1920
1921 LIRLDFSDES SEIFKNCARK VLNIIKVSPS TSSELCQMGL KFLSAFIRHT DSTLKDTALS1980
1981 YVLGRVLPDL NEPSRQGLAF NFLKALVSKH IMLPELYDIA DTTREIMVTN HSKEIRDVSR2040
2041 SVYYQFLMEY DQSKGRLEKQ FKFMVDNLQY PTESGRQSVM ELINLIITKA NPALLSKLSS2100
2101 SFFLALVNVS FNDDAPRCRE MASVLISTML PKLENKDLEI VEKYIAAWLK QVDNASFLNL2160
2161 GLRTYKVYLK SIGFEHTIEL DELAIKRIRY ILSDTSVGSE HQWDLVYSAL NTFSSYMEAT2220
2221 ESVYKHGFKD IWDGIITCLL YPHSWVRQSA ANLVHQLIAN KDKLEISLTN LEIQTIATRI2280
2281 LHQLGAPSIP ENLANVSIKT LVNISILWKE QRTPFIMDVS KQTGEDLKYT TAIDYMVTRI2340
2341 GGIIRSDEHR MDSFMSKKAC IQLLALLVQV LDEDEVIAEG EKILLPLYGY LETYYSRAVD2400
2401 EEQEELRTLS NECLKILEDK LQVSDFTKIY TAVKQTVLER RKERRSKRAI LAVNAPQISA2460
2461 DKKLRKHARS REKRKHEKDE NGYYQRRNKR KRA |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #20 Asynchronous Prep1-TiO2 Phosphopeptide enrichment, PartB | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
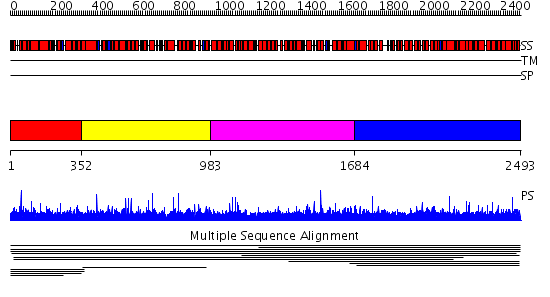
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..351] | 2.006916 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [352..982] | 1.004883 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [983..1683] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [1684..2493] | 2.008001 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [591..825] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [826..1037] | 79.283997 | No description for PF07539.3 was found. No confident structure predictions are available. | |
| 7 | View Details | [1038..1208] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1209..1317] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1318..2197] | 2.65 | No description for 2ie3A was found. | |
| 10 | View Details | [2198..2351] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [2352..2493] | 1.036996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||
| 9 |
|
||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||
| 11 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)