













| General Information: |
|
| Name(s) found: |
HSL1 /
YKL101W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1518 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular bud neck
[IDA septin ring [IDA |
| Biological Process: |
cell morphogenesis checkpoint
[TAS G2/M transition of mitotic cell cycle [IGI protein amino acid phosphorylation [IDA septin checkpoint [IGI |
| Molecular Function: |
protein kinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTGHVSKTSH VPKGRPSSLA KKAAKRAMAK VNSNPKRASG HLERVVQSVN DATKRLSQPD 60
61 STVSVATKSS KRKSRDTVGP WKLGKTLGKG SSGRVRLAKN METGQLAAIK IVPKKKAFVH 120
121 CSNNGTVPNS YSSSMVTSNV SSPSIASREH SNHSQTNPYG IEREIVIMKL ISHTNVMALF 180
181 EVWENKSELY LVLEYVDGGE LFDYLVSKGK LPEREAIHYF KQIVEGVSYC HSFNICHRDL 240
241 KPENLLLDKK NRRIKIADFG MAALELPNKL LKTSCGSPHY ASPEIVMGRP YHGGPSDVWS 300
301 CGIVLFALLT GHLPFNDDNI KKLLLKVQSG KYQMPSNLSS EARDLISKIL VIDPEKRITT 360
361 QEILKHPLIK KYDDLPVNKV LRKMRKDNMA RGKSNSDLHL LNNVSPSIVT LHSKGEIDES 420
421 ILRSLQILWH GVSRELITAK LLQKPMSEEK LFYSLLLQYK QRHSISLSSS SENKKSATES 480
481 SVNEPRIEYA SKTANNTGLR SENNDVKTLH SLEIHSEDTS TVNQNNAITG VNTEINAPVL 540
541 AQKSQFSINT LSQPESDKAE AEAVTLPPAI PIFNASSSRI FRNSYTSISS RSRRSLRLSN 600
601 SRLSLSASTS RETVHDNEMP LPQLPKSPSR YSLSRRAIHA SPSTKSIHKS LSRKNIAATV 660
661 AARRTLQNSA SKRSLYSLQS ISKRSLNLND LLVFDDPLPS KKPASENVNK SEPHSLESDS 720
721 DFEILCDQIL FGNALDRILE EEEDNEKERD TQRQRQNDTK SSADTFTISG VSTNKENEGP 780
781 EYPTKIEKNQ FNMSYKPSEN MSGLSSFPIF EKENTLSSSY LEEQKPKRAA LSDITNSFNK 840
841 MNKQEGMRIE KKIQREQLQK KNDRPSPLKP IQHQELRVNS LPNDQGKPSL SLDPRRNISQ 900
901 PVNSKVESLL QGLKFKKEPA SHWTHERGSL FMSEHVEDEK PVKASDVSIE SSYVPLTTVA 960
961 TSSRDPSVLA ESSTIQKPML SLPSSFLNTS MTFKNLSQIL ADDGDDKHLS VPQNQSRSVA1020
1021 MSHPLRKQSA KISLTPRSNL NANLSVKRNQ GSPGSYLSND LDGISDMTFA MEIPTNTFTA1080
1081 QAIQLMNNDT DNNKINTSPK ASSFTKEKVI KSAAYISKEK EPDNSDTNYI PDYTIPNTYD1140
1141 EKAINIFEDA PSDEGSLNTS SSESDSRASV HRKAVSIDTM ATTNVLTPAT NVRVSLYWNN1200
1201 NSSGIPRETT EEILSKLRLS PENPSNTHMQ KRFSSTRGSR DSNALGISQS LQSMFKDLEE1260
1261 DQDGHTSQAD ILESSMSYSK RRPSEESVNP KQRVTMLFDE EEEESKKVGG GKIKEEHTKL1320
1321 DNKISEESSQ LVLPVVEKKE NANNTENNYS KIPKPSTIKV TKDTAMESNT QTHTKKPILK1380
1381 SVQNVEVEEA PSSDKKNWFV KLFQNFSSHN NATKASKNHV TNISFDDAHM LTLNEFNKNS1440
1441 IDYQLKNLDH KFGRKVVEYD CKFVKGNFKF KIKITSTPNA SSVITVKKRS KHSNTSSNKA1500
1501 FEKFNDDVER VIRNAGRS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
BCY1 CHK1 HSL1 TPK1 TPK2 |
| View Details | Gavin AC, et al. (2006) |
|
CAM1 HSL1 RAD34 |
| View Details | Gavin AC, et al. (2002) |

|
ADE4 ASC1 BFR2 BMS1 BRX1 BUD21 CBF5 CFT2 CKA1 CKA2 CKB1 CKB2 CMS1 DBP8 DIA4 DIM1 DIP2 DIS3 ECM16 ECM29 EMG1 ENP1 ENP2 ERO1 ESF1 ESF2 FAP7 FIP1 GCN1 GLC7 HAS1 HCA4 HHF1, HHF2 HSL1 HTA2 IMP3 IMP4 KRE33 KRR1 LCP5 LTV1 MPP10 MRD1 MVD1 NAN1 NOB1 NOC2 NOC4 NOP1 NOP12 NOP14 NOP56 NOP58 NOP6 PAP1 PFS2 PNO1 PRE6 PRE9 PRP43 PTA1 PTI1 PWP2 RCL1 REF2 RML2 RNA14 ROK1 RPP2B RRP12 RRP7 RRP9 RTG2 SAM1 SCL1 SEC21 SLX9 SOF1 SRO9 TIF1, TIF2 TSR1 UTP10 UTP11 UTP13 UTP15 UTP18 UTP20 UTP21 UTP22 UTP30 UTP4 UTP6 UTP7 UTP8 UTP9 YOR059C YSH1 |
| View Details | Qiu et al. (2008) |
|
ELM1 GIN4 HSL1 SWE1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
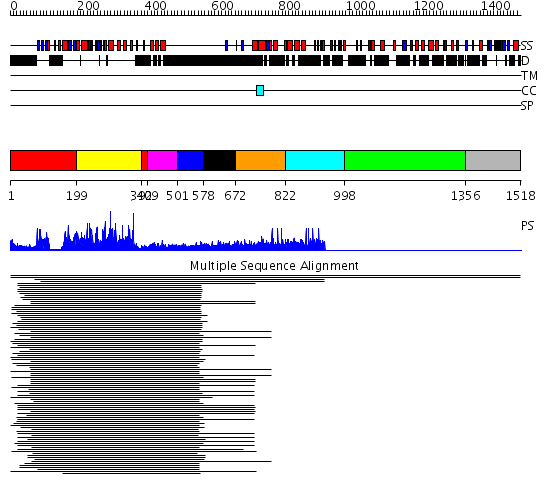
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..198] [392..408] |
579.0103 | Calmodulin-dependent protein kinase | |
| 2 | View Details | [199..391] | 579.0103 | Calmodulin-dependent protein kinase | |
| 3 | View Details | [409..500] | 11.0 | Calmodulin | |
| 4 | View Details | [501..577] | 11.0 | Calmodulin | |
| 5 | View Details | [578..671] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [672..821] | 9.74 | RBP1 | |
| 7 | View Details | [822..997] | 10.41 | EGF receptor extracellular domain; EGF receptor Cys-rich domains | |
| 8 | View Details | [998..1355] | 10.41 | EGF receptor extracellular domain; EGF receptor Cys-rich domains | |
| 9 | View Details | [1356..1518] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1354..1518] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)