













| General Information: |
|
| Name(s) found: |
NOC2 /
YOR206W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 710 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA Noc2p-Noc3p complex [IPI mitochondrion [IDA Noc1p-Noc2p complex [IPI |
| Biological Process: |
ribosomal subunit export from nucleus
[TAS ribosome biogenesis [RCA ribosome assembly [TAS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKVSKSTKK FQSKHLKHTL DQRRKEKIQK KRIQGRRGNK TDQEKADAAG TREQQQLKKS 60
61 AKEEVFKDMS VETFFEKGIE IPKENKKLKK KTTKEQSDED SSSSEEEEDM GQSMAKLAEK 120
121 DPEFYKYLEE NDKDLLDFAG TNPLDGIDSQ DEGEDAERNS NIEEKSEQME LEKEKIELSL 180
181 KLVRKWKKQL HDSPSLKLLR NIISAFKVAV NLNKEENIED YKYAITDEKA FHELMFMVLK 240
241 DVPQAIQKMA PYKIVKGART LPNGGNVSRV SSIVKSHAGS LLILLNDITN TETAALVLHS 300
301 VNELMPYLLS YRRILKELIK SIVGVWSTTR ELETQIASFA FLINTTKEFK KSMLETTLKT 360
361 TYSTFIKSCR KTNMRSMPLI NFQKNSAAEL FGIDEVLGYQ VGFEYIRQLA IHLRNTMNAT 420
421 TKKSSKINSA EAYKIVYNWQ FCHSLDFWSR VLSFACQPEK ENGSESPLRQ LIYPLVQVTL 480
481 GVIRLIPTPQ FFPLRFYLIK SLIRLSQNSG VFIPIYPLLS EILTSTAFTK APKKSPNLAA 540
541 FDFEHNIKCT QAYLNTKIYQ EGLSEQFVDL LGDYFALYCK NIAFPELVTP VIISLRRYIK 600
601 TSTNVKLNKR LSTVVEKLNQ NSTFIQEKRS DVEFGPTNKS EVSRFLNDVA WNKTPLGSYV 660
661 AVQREVKEEK ARLMRESMEE QDKERETEEA KLLNSLESDD DNEDVEMSDA |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
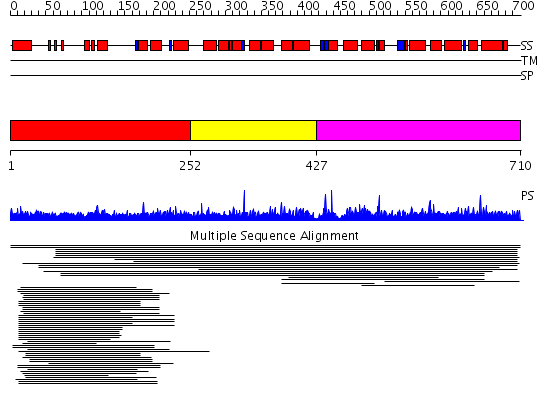
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..251] | 16.032997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [252..426] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [427..710] | 1.016967 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)